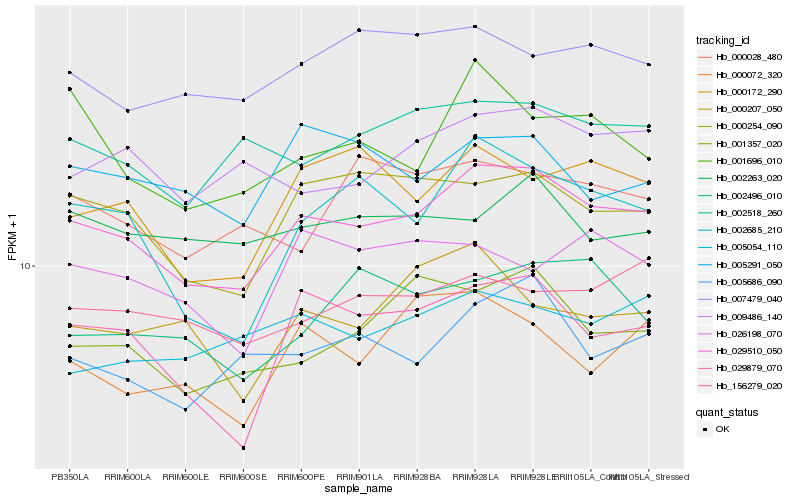
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029510_050 |
0.0 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 2 |
Hb_000254_090 |
0.0609004456 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 3 |
Hb_029879_070 |
0.0616634899 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 4 |
Hb_001357_020 |
0.0660781779 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002518_260 |
0.0662144667 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 6 |
Hb_005686_090 |
0.0665026059 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002685_210 |
0.0681531627 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000172_290 |
0.0695410096 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 9 |
Hb_009486_140 |
0.0700989675 |
- |
- |
PREDICTED: uncharacterized protein LOC105643248 [Jatropha curcas] |
| 10 |
Hb_156279_020 |
0.0714698405 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 11 |
Hb_007479_040 |
0.0719967977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001696_010 |
0.073045104 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_005291_050 |
0.0759449885 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005054_110 |
0.0759809092 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 15 |
Hb_002496_010 |
0.0764213728 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000072_320 |
0.0765081937 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 17 |
Hb_000207_050 |
0.0769010356 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002263_020 |
0.0769295011 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 19 |
Hb_000028_480 |
0.0770305272 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 20 |
Hb_026198_070 |
0.0779324915 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |