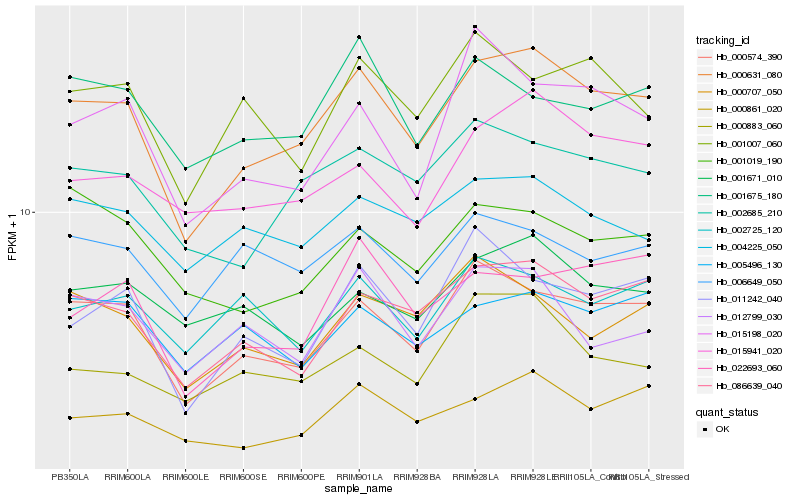
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000631_080 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 2 |
Hb_000574_390 |
0.0655477776 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 3 |
Hb_086639_040 |
0.0760727276 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 4 |
Hb_002685_210 |
0.080461288 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000861_020 |
0.0882740069 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 6 |
Hb_015941_020 |
0.0972380594 |
- |
- |
- |
| 7 |
Hb_004225_050 |
0.0988758222 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_022693_060 |
0.0989037842 |
- |
- |
- |
| 9 |
Hb_000707_050 |
0.0992041988 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011242_040 |
0.100327995 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001675_180 |
0.1014812083 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 12 |
Hb_015198_020 |
0.101665107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006649_050 |
0.1019691925 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 14 |
Hb_001671_010 |
0.1033709602 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 15 |
Hb_001007_060 |
0.1034124311 |
- |
- |
Transcriptional corepressor SEUSS [Glycine soja] |
| 16 |
Hb_005496_130 |
0.1035887552 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_012799_030 |
0.1043938285 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 18 |
Hb_002725_120 |
0.1048547947 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000883_060 |
0.1056067718 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 20 |
Hb_001019_190 |
0.1069684201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |