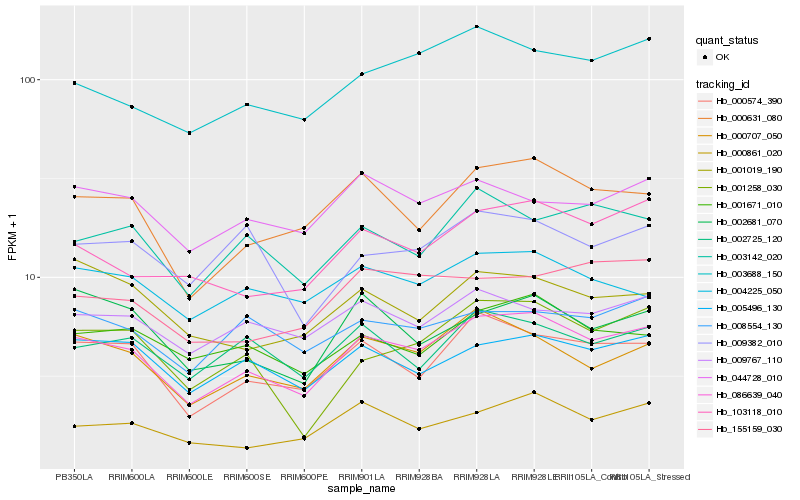
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086639_040 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 2 |
Hb_000574_390 |
0.0703695779 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 3 |
Hb_000707_050 |
0.0743888964 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000631_080 |
0.0760727276 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Jatropha curcas] |
| 5 |
Hb_005496_130 |
0.0850535907 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003688_150 |
0.0865644561 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_103118_010 |
0.0889504694 |
- |
- |
PREDICTED: uncharacterized protein LOC105643231 [Jatropha curcas] |
| 8 |
Hb_001671_010 |
0.0901485341 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 9 |
Hb_000861_020 |
0.0931960517 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 10 |
Hb_003142_020 |
0.093752255 |
- |
- |
PREDICTED: uncharacterized protein LOC105648334 [Jatropha curcas] |
| 11 |
Hb_002725_120 |
0.0949204154 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 12 |
Hb_009767_110 |
0.0956892312 |
- |
- |
- |
| 13 |
Hb_002681_070 |
0.097108181 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008554_130 |
0.0973398257 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105634317 [Jatropha curcas] |
| 15 |
Hb_044728_010 |
0.0987513739 |
- |
- |
- |
| 16 |
Hb_155159_030 |
0.1002428785 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 17 |
Hb_001258_030 |
0.1015170104 |
- |
- |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 18 |
Hb_004225_050 |
0.1020168776 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001019_190 |
0.1032681974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_009382_010 |
0.1044231762 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |