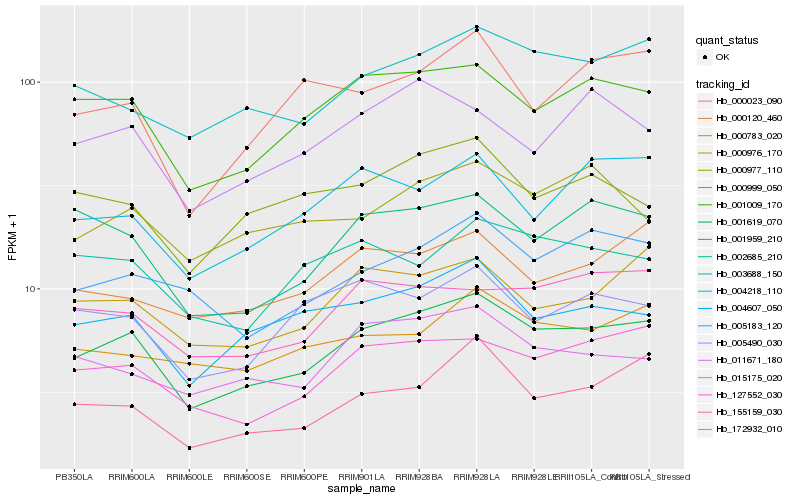
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103503585 [Cucumis melo] |
| 2 |
Hb_001009_170 |
0.0718824538 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 3 |
Hb_127552_030 |
0.0810907032 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 4 |
Hb_003688_150 |
0.0842975262 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 5 |
Hb_004607_050 |
0.0867405483 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_001959_210 |
0.0917187695 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 7 |
Hb_000976_170 |
0.0928556397 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 8 |
Hb_005183_120 |
0.0930832885 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 9 |
Hb_155159_030 |
0.0952731994 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 10 |
Hb_015175_020 |
0.0956759231 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |
| 11 |
Hb_172932_010 |
0.0963544876 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000999_050 |
0.09988411 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004218_110 |
0.100850712 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 14 |
Hb_002685_210 |
0.1015026391 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005490_030 |
0.101645112 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000783_020 |
0.1017378693 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase SDP6, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000120_460 |
0.1023523232 |
- |
- |
PREDICTED: thioredoxin reductase NTRB-like [Jatropha curcas] |
| 18 |
Hb_000977_110 |
0.1036432081 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000023_090 |
0.1044915541 |
- |
- |
mak, putative [Ricinus communis] |
| 20 |
Hb_011671_180 |
0.1046583776 |
- |
- |
PREDICTED: probable quinone oxidoreductase [Jatropha curcas] |