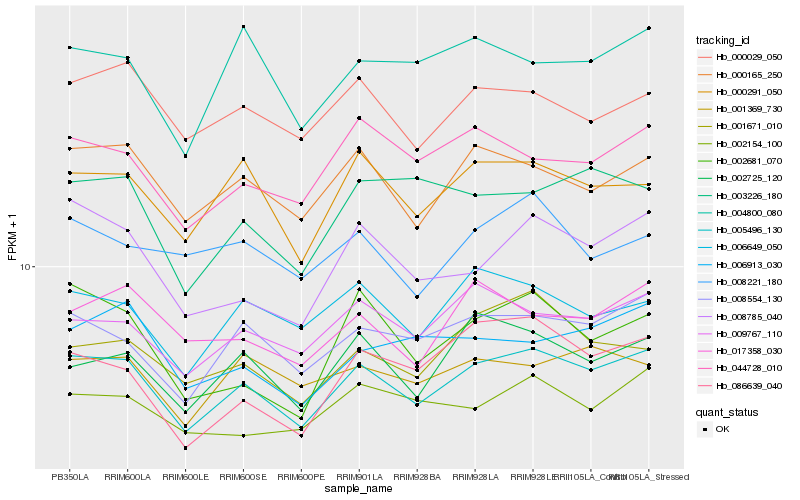
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005496_130 |
0.0 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_008554_130 |
0.0605009463 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105634317 [Jatropha curcas] |
| 3 |
Hb_000165_250 |
0.0620235685 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 4 |
Hb_000291_050 |
0.0669112915 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009767_110 |
0.0675886848 |
- |
- |
- |
| 6 |
Hb_002725_120 |
0.0719919063 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000029_050 |
0.0726842045 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 8 |
Hb_008785_040 |
0.0727730454 |
- |
- |
PREDICTED: DNA repair protein UVH3 [Jatropha curcas] |
| 9 |
Hb_044728_010 |
0.0728176582 |
- |
- |
- |
| 10 |
Hb_006649_050 |
0.0730062572 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 11 |
Hb_017358_030 |
0.0789513963 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001671_010 |
0.0794860593 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 13 |
Hb_001369_730 |
0.0804785352 |
- |
- |
F5O11.10, putative isoform 1 [Theobroma cacao] |
| 14 |
Hb_004800_080 |
0.081167088 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 15 |
Hb_002154_100 |
0.0812324464 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 16 |
Hb_006913_030 |
0.0833041072 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 17 |
Hb_003226_180 |
0.0843622869 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 18 |
Hb_086639_040 |
0.0850535907 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 19 |
Hb_002681_070 |
0.0855489713 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008221_180 |
0.0859901339 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |