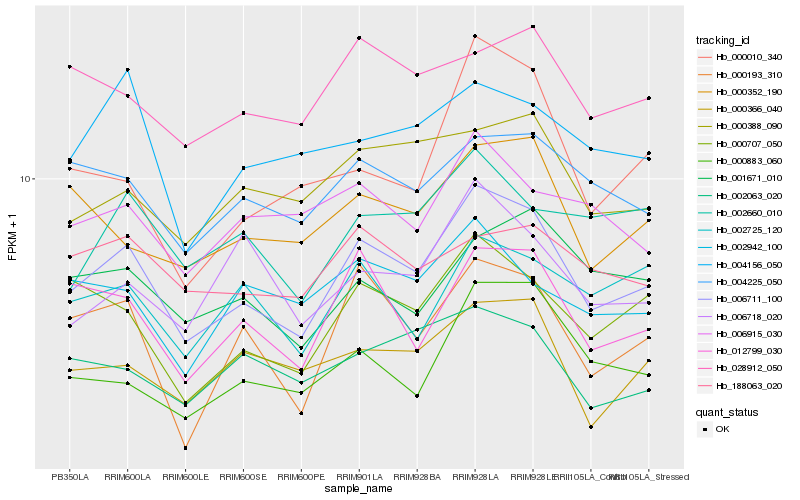
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006711_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637078 [Jatropha curcas] |
| 2 |
Hb_012799_030 |
0.0844260741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 3 |
Hb_000707_050 |
0.0846634855 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004225_050 |
0.0922598646 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000388_090 |
0.0926279746 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_000883_060 |
0.0941119674 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 7 |
Hb_000352_190 |
0.0942035995 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 41 [Jatropha curcas] |
| 8 |
Hb_000193_310 |
0.0943622807 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 9 |
Hb_000366_040 |
0.0943919268 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 10 |
Hb_001671_010 |
0.0973574004 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 11 |
Hb_028912_050 |
0.0977543891 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002725_120 |
0.0985522673 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002660_010 |
0.0987213043 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18-like [Jatropha curcas] |
| 14 |
Hb_002063_020 |
0.0987820519 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 15 |
Hb_006915_030 |
0.1013025134 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 8 [Jatropha curcas] |
| 16 |
Hb_004156_050 |
0.1015230622 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 17 |
Hb_188063_020 |
0.1015247695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002942_100 |
0.1015252914 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |
| 19 |
Hb_006718_020 |
0.102313222 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000010_340 |
0.1033204109 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |