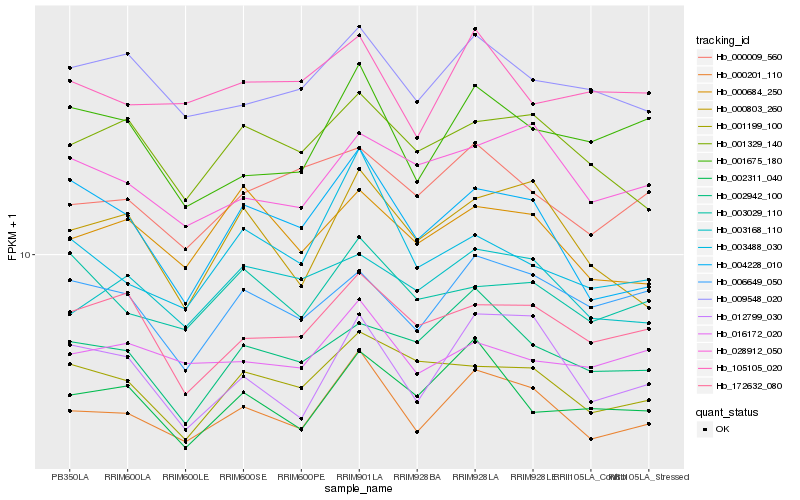
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000201_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_21690 [Jatropha curcas] |
| 2 |
Hb_012799_030 |
0.0756897742 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g55630-like [Jatropha curcas] |
| 3 |
Hb_009548_020 |
0.0783911449 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_001329_140 |
0.0831451641 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 5 |
Hb_028912_050 |
0.0839008171 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006649_050 |
0.0844830369 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 7 |
Hb_001675_180 |
0.0845761925 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 8 |
Hb_002311_040 |
0.0846579938 |
- |
- |
hypothetical protein JCGZ_18966 [Jatropha curcas] |
| 9 |
Hb_003168_110 |
0.0850149351 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 10 |
Hb_000009_560 |
0.0850209364 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_000803_260 |
0.085087233 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 12 |
Hb_016172_020 |
0.0852944838 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_172632_080 |
0.0861779213 |
- |
- |
PREDICTED: uncharacterized protein LOC105141138 isoform X1 [Populus euphratica] |
| 14 |
Hb_000684_250 |
0.0862175644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642172 [Jatropha curcas] |
| 15 |
Hb_004228_010 |
0.086702879 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA [Jatropha curcas] |
| 16 |
Hb_105105_020 |
0.0877226841 |
- |
- |
vacuolar protein sorting vps41, putative [Ricinus communis] |
| 17 |
Hb_001199_100 |
0.0879354459 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 18 |
Hb_003488_030 |
0.0882806657 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 19 |
Hb_003029_110 |
0.0898882169 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002942_100 |
0.0918250045 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |