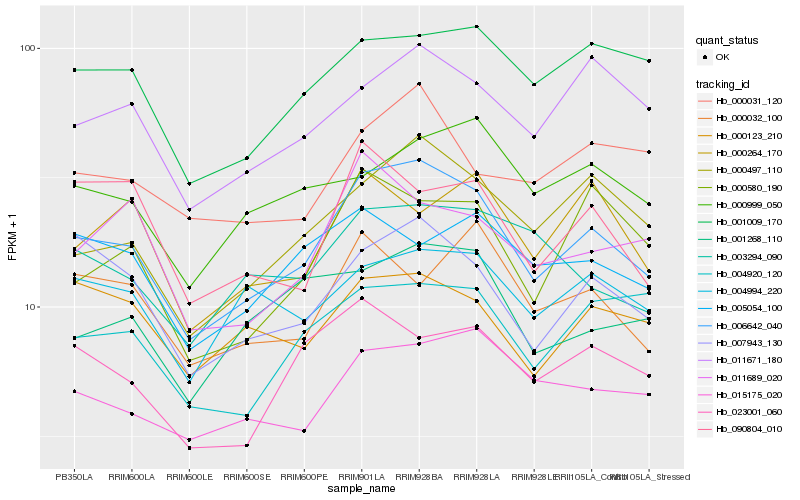
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006642_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 2 |
Hb_000497_110 |
0.0920351416 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 3 |
Hb_007943_130 |
0.0958882739 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 4 |
Hb_015175_020 |
0.1055980715 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |
| 5 |
Hb_011671_180 |
0.1061208728 |
- |
- |
PREDICTED: probable quinone oxidoreductase [Jatropha curcas] |
| 6 |
Hb_001009_170 |
0.1083427525 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_023001_060 |
0.109316676 |
- |
- |
PREDICTED: importin-11 [Jatropha curcas] |
| 8 |
Hb_004994_220 |
0.1114727951 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000580_190 |
0.1115219392 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 10 |
Hb_001268_110 |
0.1128191208 |
- |
- |
PREDICTED: uncharacterized protein LOC105630183 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003294_090 |
0.1144302418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000999_050 |
0.1156964345 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004920_120 |
0.1173048129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000123_210 |
0.1175479283 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011689_020 |
0.1179231283 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_005054_100 |
0.1187951457 |
- |
- |
PREDICTED: uncharacterized protein At2g39920 [Jatropha curcas] |
| 17 |
Hb_000031_120 |
0.119507802 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 18 |
Hb_000264_170 |
0.1201051281 |
- |
- |
PREDICTED: uncharacterized protein LOC105649522 [Jatropha curcas] |
| 19 |
Hb_090804_010 |
0.1201175613 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000032_100 |
0.1207234086 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Vitis vinifera] |