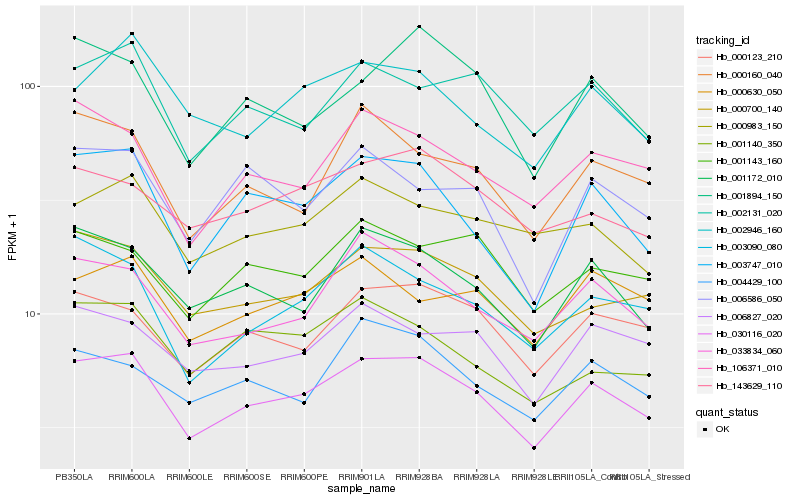
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030116_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s13630g [Populus trichocarpa] |
| 2 |
Hb_003747_010 |
0.0679125998 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 3 |
Hb_001172_010 |
0.0725181938 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 4 |
Hb_000700_140 |
0.0728966795 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 5 |
Hb_000123_210 |
0.0739409824 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 6 |
Hb_033834_060 |
0.0739741551 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_106371_010 |
0.0811102845 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_002131_020 |
0.0814959431 |
transcription factor |
TF Family: IWS1 |
PREDICTED: protein IWS1 homolog [Jatropha curcas] |
| 9 |
Hb_006827_020 |
0.0815511228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003090_080 |
0.0817306825 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein B [Jatropha curcas] |
| 11 |
Hb_000160_040 |
0.0847384068 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 12 |
Hb_143629_110 |
0.0851400317 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 13 |
Hb_001894_150 |
0.0851476246 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 14 |
Hb_002946_160 |
0.0871374785 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 15 |
Hb_000630_050 |
0.0880139957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004429_100 |
0.0880311469 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_001143_160 |
0.0882633325 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000983_150 |
0.0893373563 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 19 |
Hb_006586_050 |
0.0903989491 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 20 |
Hb_001140_350 |
0.0908273656 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |