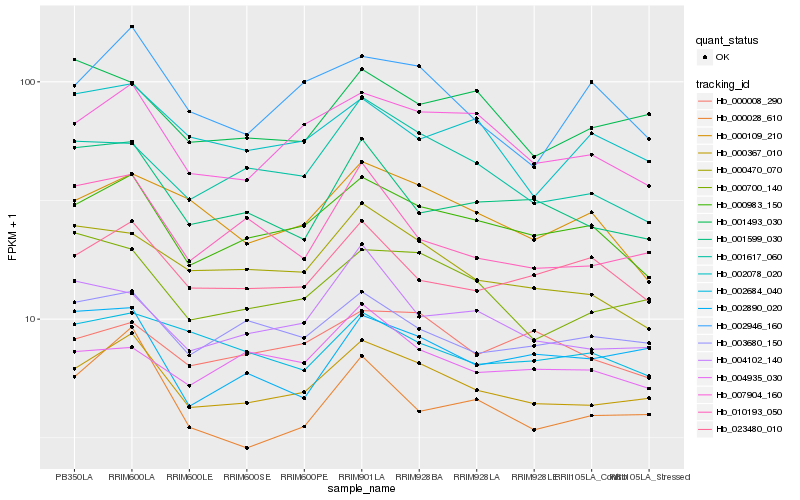
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_010 |
0.0 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 2 |
Hb_000983_150 |
0.0582078697 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 3 |
Hb_007904_160 |
0.0650550627 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 4 |
Hb_003680_150 |
0.0687029329 |
- |
- |
srpk, putative [Ricinus communis] |
| 5 |
Hb_023480_010 |
0.0752371894 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000700_140 |
0.0781763792 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 7 |
Hb_001617_060 |
0.0816704575 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 8 |
Hb_000470_070 |
0.0826939794 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 9 |
Hb_000008_290 |
0.082829426 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 10 |
Hb_002684_040 |
0.0845720796 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004102_140 |
0.0846234798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002890_020 |
0.0855080924 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004935_030 |
0.0862633478 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 14 |
Hb_002078_020 |
0.0865734616 |
- |
- |
PREDICTED: proteasome subunit beta type-5 [Jatropha curcas] |
| 15 |
Hb_000109_210 |
0.0866654836 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_010193_050 |
0.0866816047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002946_160 |
0.0872819213 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 18 |
Hb_001599_030 |
0.0876255527 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_000028_610 |
0.0880144923 |
- |
- |
PREDICTED: uncharacterized protein LOC105644291 [Jatropha curcas] |
| 20 |
Hb_001493_030 |
0.0886062298 |
- |
- |
hypothetical protein MIMGU_mgv1a005351mg [Erythranthe guttata] |