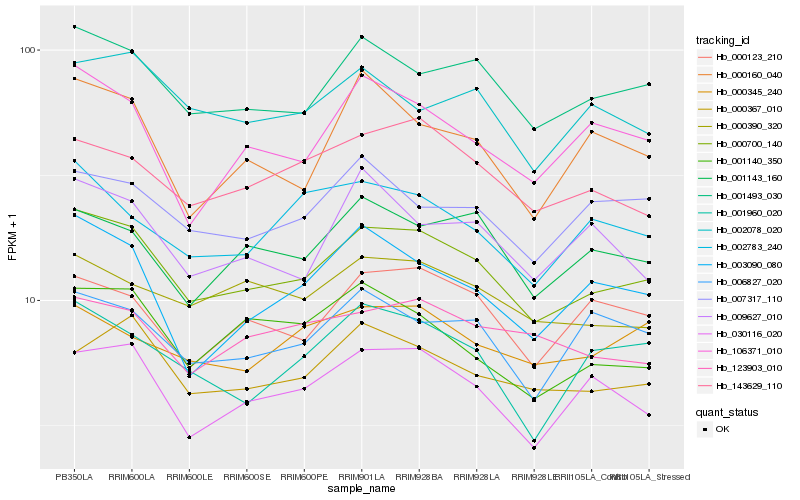
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_140 |
0.0 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 2 |
Hb_001493_030 |
0.0548180974 |
- |
- |
hypothetical protein MIMGU_mgv1a005351mg [Erythranthe guttata] |
| 3 |
Hb_030116_020 |
0.0728966795 |
- |
- |
hypothetical protein POPTR_0016s13630g [Populus trichocarpa] |
| 4 |
Hb_143629_110 |
0.0756101684 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 5 |
Hb_000123_210 |
0.0776341408 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000367_010 |
0.0781763792 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 7 |
Hb_106371_010 |
0.0790736839 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_001143_160 |
0.0794039508 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006827_020 |
0.0798933724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000390_320 |
0.0801683802 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 11 |
Hb_007317_110 |
0.0810081244 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C660.15-like [Jatropha curcas] |
| 12 |
Hb_001960_020 |
0.0812781534 |
- |
- |
PREDICTED: mitochondrial arginine transporter BAC2 [Jatropha curcas] |
| 13 |
Hb_003090_080 |
0.0820683076 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein B [Jatropha curcas] |
| 14 |
Hb_002078_020 |
0.0823486255 |
- |
- |
PREDICTED: proteasome subunit beta type-5 [Jatropha curcas] |
| 15 |
Hb_002783_240 |
0.0827586172 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 16 |
Hb_001140_350 |
0.0828065929 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 17 |
Hb_123903_010 |
0.0855335645 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000160_040 |
0.0864218932 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 19 |
Hb_000345_240 |
0.0870680689 |
- |
- |
PREDICTED: cyclin-H1-1 [Jatropha curcas] |
| 20 |
Hb_009627_010 |
0.088210892 |
- |
- |
Protein dom-3, putative [Ricinus communis] |