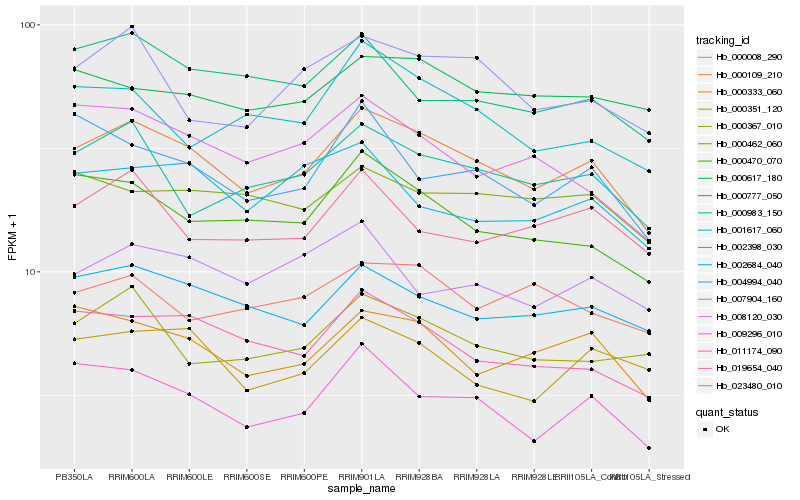
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_210 |
0.0 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002684_040 |
0.0703412373 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000470_070 |
0.0724518929 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 4 |
Hb_000333_060 |
0.0733535715 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 5 |
Hb_019654_040 |
0.0738998254 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000983_150 |
0.0761460741 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 7 |
Hb_011174_090 |
0.078886842 |
- |
- |
PREDICTED: fidgetin-like protein 1 [Jatropha curcas] |
| 8 |
Hb_007904_160 |
0.0827941065 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 9 |
Hb_000777_050 |
0.0847688236 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 10 |
Hb_009296_010 |
0.0849287627 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 11 |
Hb_004994_040 |
0.085257856 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 12 |
Hb_000351_120 |
0.0854003046 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 13 |
Hb_001617_060 |
0.0856439301 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 14 |
Hb_023480_010 |
0.0861153137 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_000367_010 |
0.0866654836 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 16 |
Hb_000462_060 |
0.0871280545 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000617_180 |
0.087550185 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 18 |
Hb_008120_030 |
0.0878311745 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 19 |
Hb_002398_030 |
0.0883046713 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000008_290 |
0.0904549558 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |