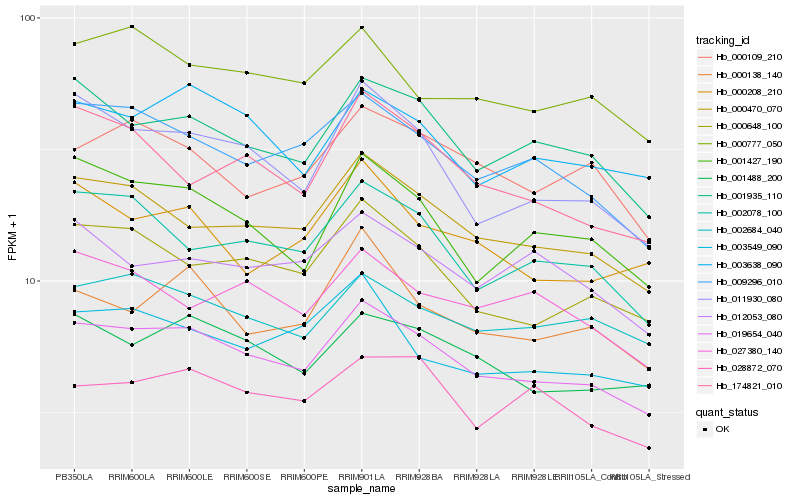
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019654_040 |
0.0 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000470_070 |
0.0512319816 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 3 |
Hb_009296_010 |
0.0565215601 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 4 |
Hb_001935_110 |
0.0573921651 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 5 |
Hb_000648_100 |
0.0591607258 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 6 |
Hb_001488_200 |
0.0609885829 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_011930_080 |
0.0630143569 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 8 |
Hb_003638_090 |
0.064610316 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 9 |
Hb_002078_100 |
0.0647103396 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 10 |
Hb_001427_190 |
0.0669390794 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 11 |
Hb_002684_040 |
0.0676450549 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000777_050 |
0.0697927817 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 13 |
Hb_000109_210 |
0.0738998254 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000138_140 |
0.0761073262 |
- |
- |
hypothetical protein B456_012G1014001, partial [Gossypium raimondii] |
| 15 |
Hb_003549_090 |
0.0762674065 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_028872_070 |
0.0762793873 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_174821_010 |
0.0764831688 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 18 |
Hb_027380_140 |
0.0779455881 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 19 |
Hb_012053_080 |
0.0784757772 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 20 |
Hb_000208_210 |
0.0785281171 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |