| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
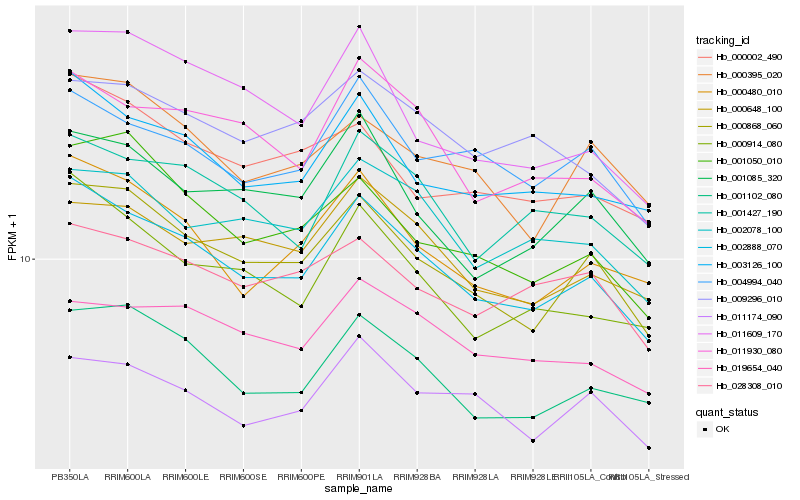
Hb_002888_070 |
0.0 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 2 |
Hb_000868_060 |
0.0521189005 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 3 |
Hb_003126_100 |
0.0577101606 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 4 |
Hb_000480_010 |
0.0634364994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000914_080 |
0.0681274696 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_001102_080 |
0.0693916265 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 7 |
Hb_011930_080 |
0.0701538778 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 8 |
Hb_001427_190 |
0.0723238367 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 9 |
Hb_002078_100 |
0.0786306606 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 10 |
Hb_004994_040 |
0.0793434987 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 11 |
Hb_000648_100 |
0.080745749 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 12 |
Hb_001085_320 |
0.0813571503 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_000395_020 |
0.0815835797 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 14 |
Hb_000002_490 |
0.0822017525 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 15 |
Hb_011609_170 |
0.0829755803 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 16 |
Hb_011174_090 |
0.0830234257 |
- |
- |
PREDICTED: fidgetin-like protein 1 [Jatropha curcas] |
| 17 |
Hb_001050_010 |
0.0835589223 |
- |
- |
Advillin [Gossypium arboreum] |
| 18 |
Hb_019654_040 |
0.0858590507 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 19 |
Hb_028308_010 |
0.0863591812 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009296_010 |
0.0866321976 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |