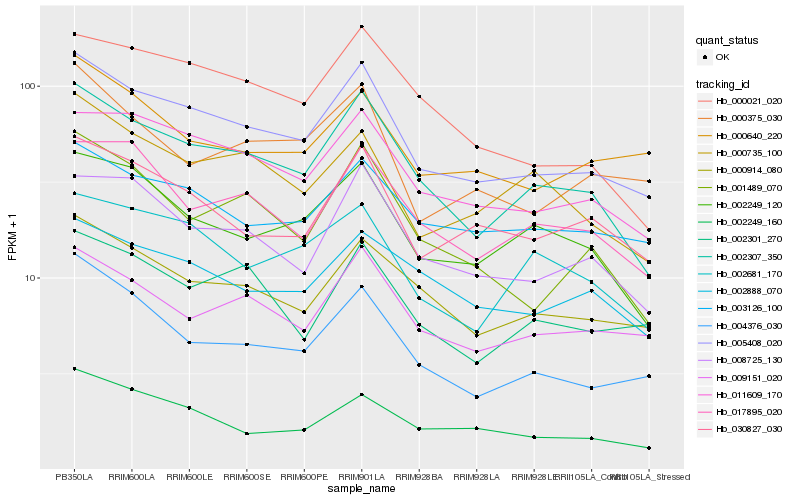
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005408_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002307_350 |
0.063515994 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 3 |
Hb_011609_170 |
0.0692568218 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 4 |
Hb_000914_080 |
0.0754654076 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_008725_130 |
0.0790845009 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 6 |
Hb_009151_020 |
0.0818935033 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 7 |
Hb_030827_030 |
0.0834763183 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 8 |
Hb_004376_030 |
0.0844699359 |
transcription factor |
TF Family: mTERF |
hypothetical protein PHAVU_005G060900g [Phaseolus vulgaris] |
| 9 |
Hb_003126_100 |
0.0848849025 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 10 |
Hb_002249_160 |
0.0901075733 |
- |
- |
- |
| 11 |
Hb_002249_120 |
0.0905387602 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 12 |
Hb_017895_020 |
0.0928057458 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 13 |
Hb_000375_030 |
0.0937460203 |
- |
- |
- |
| 14 |
Hb_002888_070 |
0.0942686146 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 15 |
Hb_000021_020 |
0.0947762448 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 16 |
Hb_000640_220 |
0.0949496633 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 17 |
Hb_002301_270 |
0.0950183231 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 18 |
Hb_001489_070 |
0.0966128023 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 19 |
Hb_000735_100 |
0.0970336502 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 20 |
Hb_002681_170 |
0.0982330356 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |