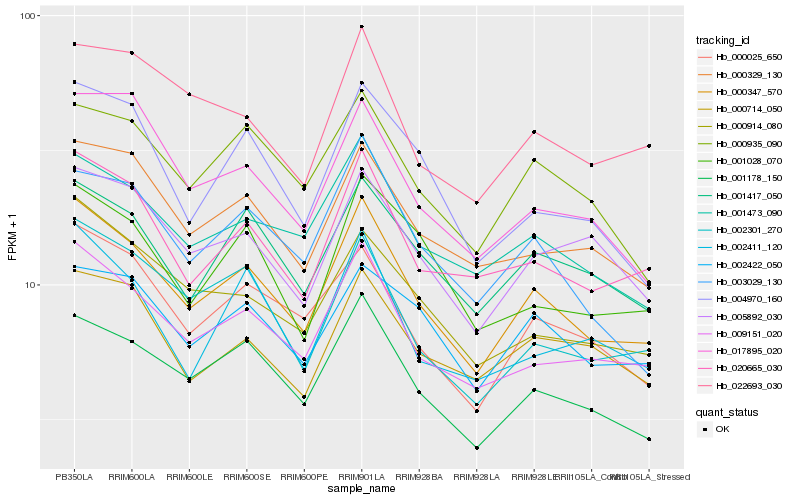
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_570 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 2 |
Hb_001178_150 |
0.0551107487 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 3 |
Hb_009151_020 |
0.0552890795 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 4 |
Hb_001473_090 |
0.0672177483 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 5 |
Hb_020665_030 |
0.067452234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000025_650 |
0.0710827848 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 7 |
Hb_001417_050 |
0.0720112859 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 8 |
Hb_000329_130 |
0.0733438411 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 9 |
Hb_000914_080 |
0.0757013453 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_002301_270 |
0.0760705372 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 11 |
Hb_000935_090 |
0.0791712289 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 12 |
Hb_002422_050 |
0.07973013 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005892_030 |
0.0801041673 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 14 |
Hb_017895_020 |
0.0811684224 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 15 |
Hb_022693_030 |
0.0836963873 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 16 |
Hb_003029_130 |
0.0838650191 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 17 |
Hb_004970_160 |
0.0843122868 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000714_050 |
0.0845527086 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 19 |
Hb_001028_070 |
0.0846388617 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002411_120 |
0.0869552086 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |