| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
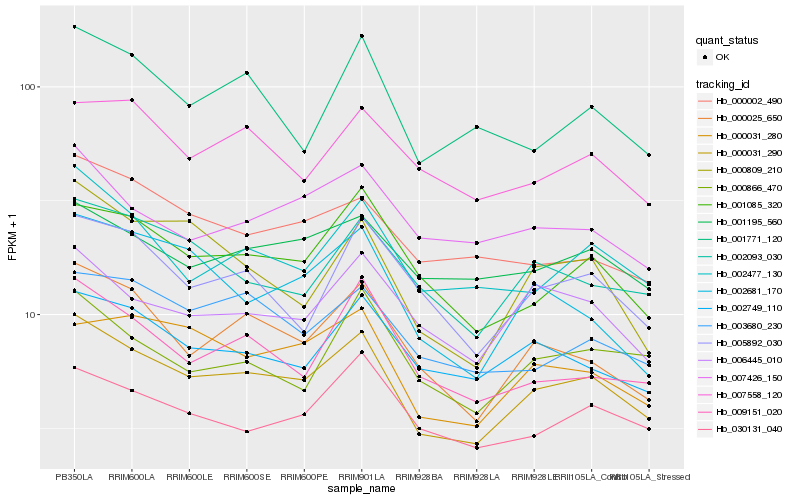
Hb_000031_290 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s28540g [Populus trichocarpa] |
| 2 |
Hb_000025_650 |
0.0762332108 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 3 |
Hb_001085_320 |
0.0829995705 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_002477_130 |
0.0851871109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000031_280 |
0.0868649994 |
- |
- |
- |
| 6 |
Hb_002749_110 |
0.0892623015 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002093_030 |
0.0905928639 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 8 |
Hb_003680_230 |
0.0906282288 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 9 |
Hb_006445_010 |
0.0914389215 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 10 |
Hb_009151_020 |
0.0926280462 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 11 |
Hb_005892_030 |
0.0943410258 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 12 |
Hb_030131_040 |
0.0947553619 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 13 |
Hb_000809_210 |
0.0960136344 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 14 |
Hb_001195_560 |
0.0960976398 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002681_170 |
0.0969829956 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 16 |
Hb_000002_490 |
0.0978752565 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 17 |
Hb_001771_120 |
0.0980592411 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 18 |
Hb_007558_120 |
0.0981042571 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 19 |
Hb_000866_470 |
0.0997894073 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 20 |
Hb_007426_150 |
0.1001651892 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |