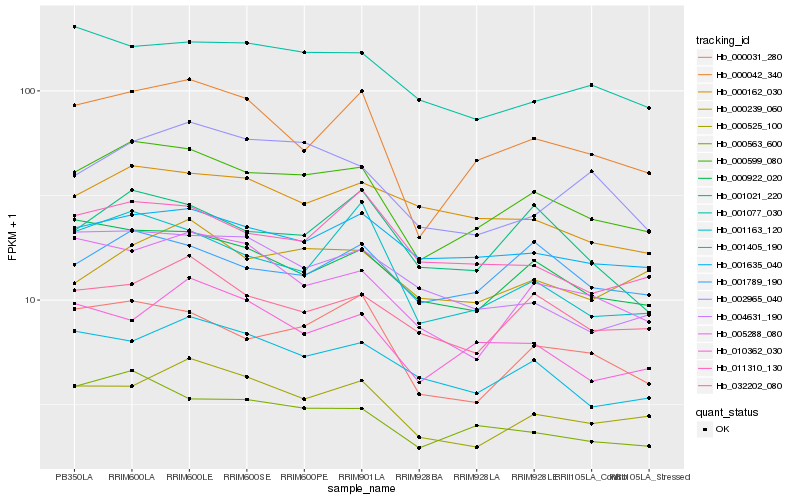
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_080 |
0.0 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 2 |
Hb_000042_340 |
0.0740702799 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 3 |
Hb_000563_600 |
0.0762021898 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 4 |
Hb_000922_020 |
0.0770018971 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000031_280 |
0.0780362881 |
- |
- |
- |
| 6 |
Hb_000525_100 |
0.0820294698 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 7 |
Hb_032202_080 |
0.0831143717 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001635_040 |
0.0838818567 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 9 |
Hb_001789_190 |
0.0850703419 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 10 |
Hb_001405_190 |
0.0891523435 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001077_030 |
0.0918333237 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001021_220 |
0.0931357213 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005288_080 |
0.0933240094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011310_130 |
0.0935172173 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_001163_120 |
0.0948353857 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_000239_060 |
0.0956716306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000162_030 |
0.0959244156 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_010362_030 |
0.0961035898 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 19 |
Hb_004631_190 |
0.0973116456 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_002965_040 |
0.09772209 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |