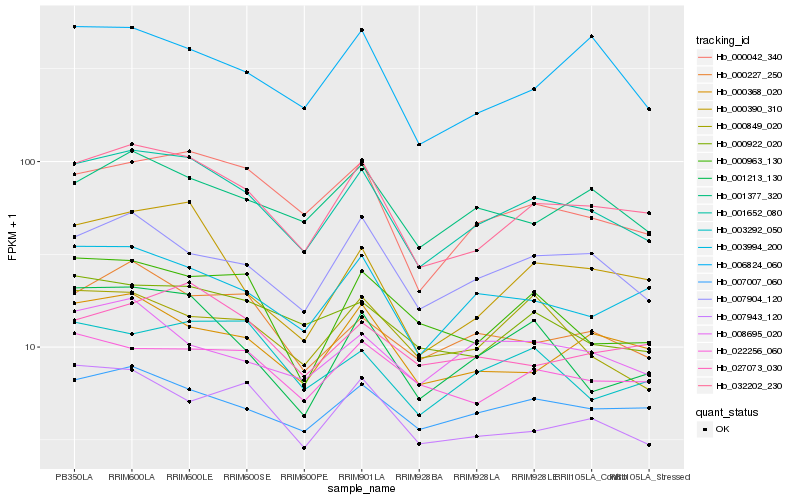
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001652_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 2 |
Hb_032202_230 |
0.0519262592 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 3 |
Hb_000042_340 |
0.0775694353 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 4 |
Hb_007904_120 |
0.0824445497 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003994_200 |
0.0876138777 |
- |
- |
PREDICTED: uncharacterized protein LOC105646142 [Jatropha curcas] |
| 6 |
Hb_007007_060 |
0.0888933746 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000227_250 |
0.0891191708 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 8 |
Hb_001377_320 |
0.0902418728 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 9 |
Hb_001213_130 |
0.0922382289 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000922_020 |
0.0927597635 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_000368_020 |
0.0950337482 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000390_310 |
0.0980370318 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007943_120 |
0.098350815 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 14 |
Hb_006824_060 |
0.0991531719 |
- |
- |
PREDICTED: uncharacterized protein LOC105650737 [Jatropha curcas] |
| 15 |
Hb_000963_130 |
0.1000419326 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_022256_060 |
0.1012484476 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 17 |
Hb_027073_030 |
0.1019338907 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 18 |
Hb_000849_020 |
0.1027845736 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008695_020 |
0.1033870569 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 20 |
Hb_003292_050 |
0.1037563041 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |