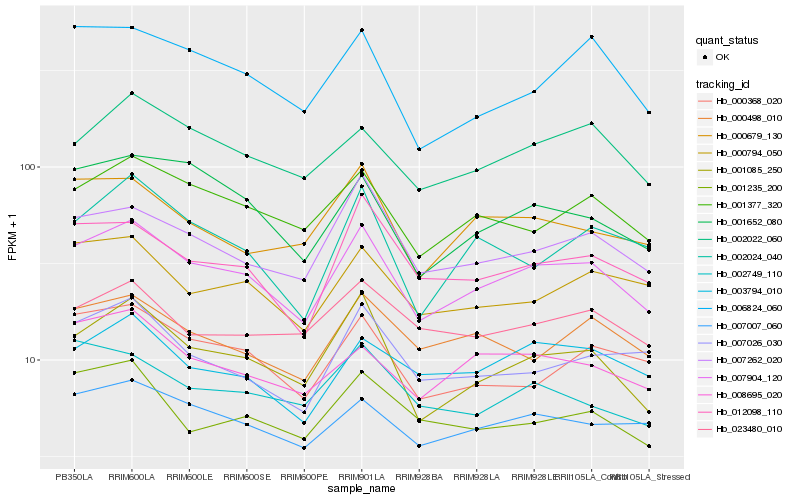
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_120 |
0.0 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001085_250 |
0.0652058615 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL isoform X2 [Jatropha curcas] |
| 3 |
Hb_001377_320 |
0.0695042916 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 4 |
Hb_007262_020 |
0.073530133 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 5 |
Hb_007007_060 |
0.0765393722 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_012098_110 |
0.0767087338 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 7 |
Hb_002022_060 |
0.0767787346 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 8 |
Hb_008695_020 |
0.0822825382 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 9 |
Hb_000794_050 |
0.082286086 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 10 |
Hb_001652_080 |
0.0824445497 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 11 |
Hb_000368_020 |
0.0831129665 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000498_010 |
0.0837211339 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_003794_010 |
0.0844362555 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 14 |
Hb_000679_130 |
0.0845410666 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_006824_060 |
0.0858262594 |
- |
- |
PREDICTED: uncharacterized protein LOC105650737 [Jatropha curcas] |
| 16 |
Hb_007026_030 |
0.0889544917 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 17 |
Hb_001235_200 |
0.0890996398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002024_040 |
0.0910221947 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 19 |
Hb_023480_010 |
0.0919956371 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_002749_110 |
0.092980917 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |