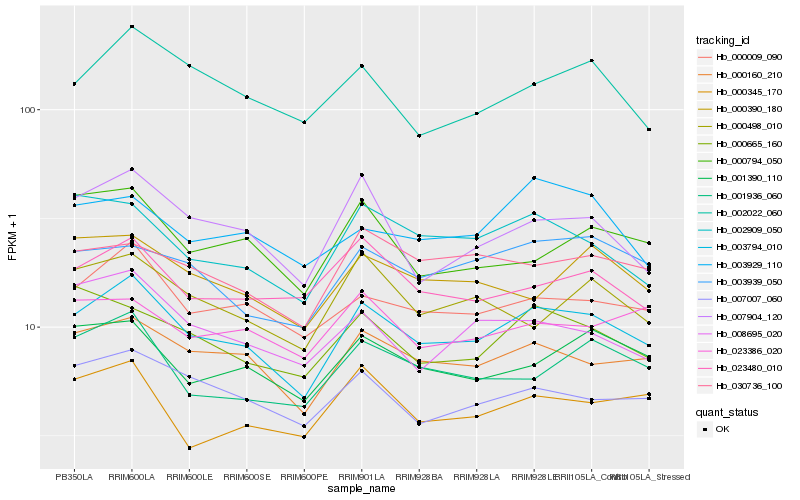
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003794_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 2 |
Hb_000160_210 |
0.0625298397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000009_090 |
0.068574011 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 4 |
Hb_007007_060 |
0.0794208759 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001390_110 |
0.0832813322 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 6 |
Hb_007904_120 |
0.0844362555 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002909_050 |
0.0850602529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001936_060 |
0.0856870836 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 9 |
Hb_030736_100 |
0.0864330685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003939_050 |
0.0868811615 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_002022_060 |
0.0871798762 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 12 |
Hb_023480_010 |
0.0886628987 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000390_180 |
0.0897883046 |
- |
- |
PREDICTED: uncharacterized protein LOC101291673 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_008695_020 |
0.0901691483 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 15 |
Hb_000665_160 |
0.0909293034 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000498_010 |
0.0923213162 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000345_170 |
0.0940300224 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000794_050 |
0.0948588325 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 19 |
Hb_003929_110 |
0.0955384855 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_023386_020 |
0.0976875299 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |