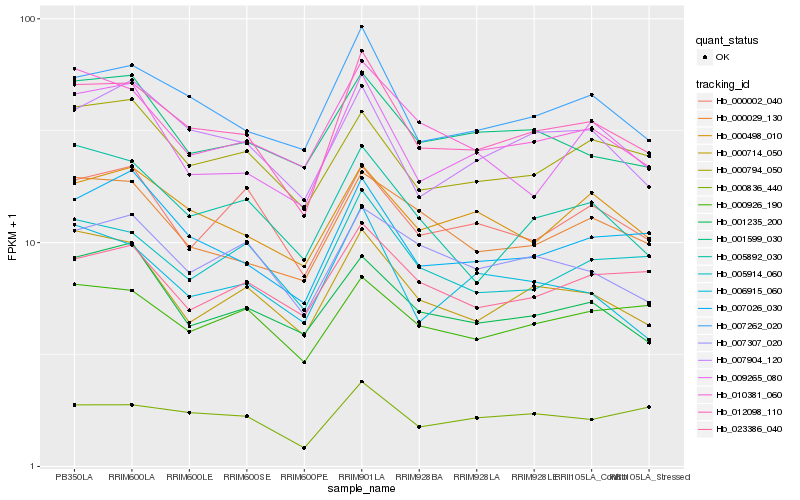
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012098_110 |
0.0 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 2 |
Hb_007262_020 |
0.0584371996 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 3 |
Hb_007026_030 |
0.0749911054 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 4 |
Hb_007904_120 |
0.0767087338 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010381_060 |
0.0771048814 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 6 |
Hb_000714_050 |
0.0784023283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 7 |
Hb_000498_010 |
0.0790213847 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_005914_060 |
0.0796343467 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 9 |
Hb_000794_050 |
0.083204322 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 10 |
Hb_000836_440 |
0.0832535219 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000029_130 |
0.0837444807 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001599_030 |
0.0851756795 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 13 |
Hb_000926_190 |
0.0868801066 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_007307_020 |
0.0871411463 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_005892_030 |
0.0882515109 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 16 |
Hb_009265_080 |
0.08893167 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 17 |
Hb_000002_040 |
0.089423774 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 18 |
Hb_023386_040 |
0.0899597221 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 19 |
Hb_001235_200 |
0.0899686832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006915_060 |
0.0905298728 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |