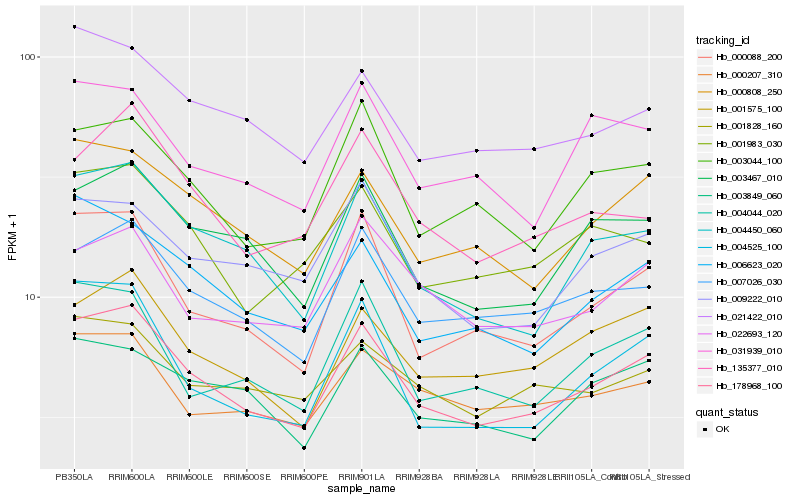
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178968_100 |
0.0 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 2 |
Hb_004450_060 |
0.0677712205 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007026_030 |
0.0695034813 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 4 |
Hb_000088_200 |
0.0705512141 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_000808_250 |
0.0709329254 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 6 |
Hb_003467_010 |
0.0733458774 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 7 |
Hb_006623_020 |
0.0776301201 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001575_100 |
0.0781081488 |
- |
- |
PREDICTED: NADPH:adrenodoxin oxidoreductase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_003044_100 |
0.0800215283 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 10 |
Hb_001983_030 |
0.0823364922 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 11 |
Hb_021422_010 |
0.0834126891 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_009222_010 |
0.0843154437 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_135377_010 |
0.0846878596 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 14 |
Hb_003849_060 |
0.0849216627 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 15 |
Hb_004044_020 |
0.0863032449 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004525_100 |
0.0870801821 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 17 |
Hb_000207_310 |
0.0894434381 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 18 |
Hb_031939_010 |
0.089693105 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001828_160 |
0.0899267592 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 20 |
Hb_022693_120 |
0.0916522604 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |