| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
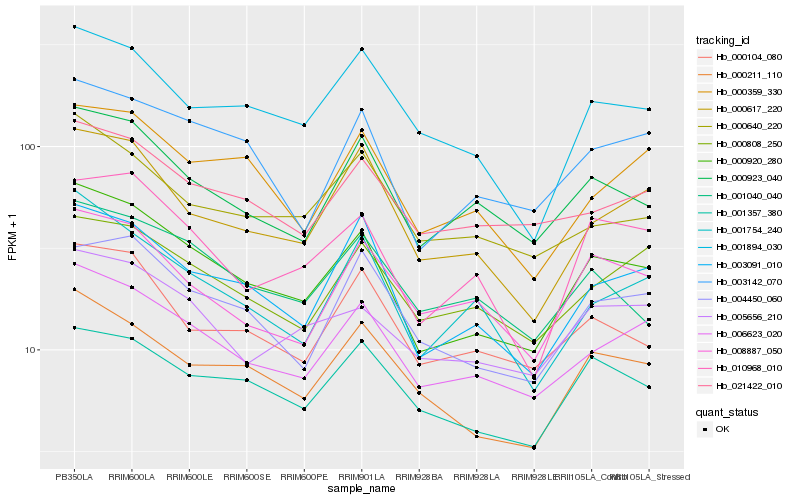
Hb_000920_280 |
0.0 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 2 |
Hb_006623_020 |
0.0738067997 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003091_010 |
0.0778361811 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 4 |
Hb_000211_110 |
0.0813362797 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_000923_040 |
0.0855168202 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 6 |
Hb_003142_070 |
0.0859141838 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 7 |
Hb_010968_010 |
0.0902203274 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 8 |
Hb_000617_220 |
0.0912263475 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 9 |
Hb_004450_060 |
0.09280658 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001754_240 |
0.0934057799 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 11 |
Hb_005656_210 |
0.0935708517 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 12 |
Hb_000104_080 |
0.0938585286 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 13 |
Hb_001357_380 |
0.0939904831 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_001894_030 |
0.0944654286 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 15 |
Hb_001040_040 |
0.1000029536 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 16 |
Hb_000359_330 |
0.1006565543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_021422_010 |
0.1013840992 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000640_220 |
0.1022218558 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 19 |
Hb_000808_250 |
0.1026266343 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 20 |
Hb_008887_050 |
0.1042628472 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |