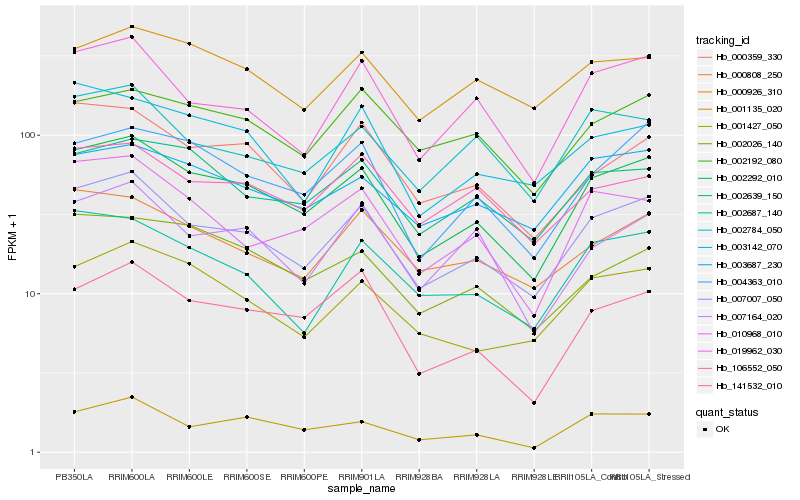
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002292_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 2 |
Hb_007007_050 |
0.0448973324 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 3 |
Hb_004363_010 |
0.0757325457 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 4 |
Hb_141532_010 |
0.0820623324 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 5 |
Hb_001135_020 |
0.0831858499 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 6 |
Hb_002639_150 |
0.0860079502 |
- |
- |
- |
| 7 |
Hb_003687_230 |
0.0864020869 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 8 |
Hb_000359_330 |
0.0880640643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001427_050 |
0.0886115237 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 10 |
Hb_010968_010 |
0.0893493721 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 11 |
Hb_002026_140 |
0.0899742529 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 12 |
Hb_019962_030 |
0.093302501 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 13 |
Hb_000808_250 |
0.0937153141 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 14 |
Hb_002687_140 |
0.0956460765 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 15 |
Hb_106552_050 |
0.098099944 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 16 |
Hb_007164_020 |
0.0991069285 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 17 |
Hb_002192_080 |
0.0998643194 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 18 |
Hb_002784_050 |
0.1006271621 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 [Jatropha curcas] |
| 19 |
Hb_003142_070 |
0.1016498205 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 20 |
Hb_000926_310 |
0.1018570922 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |