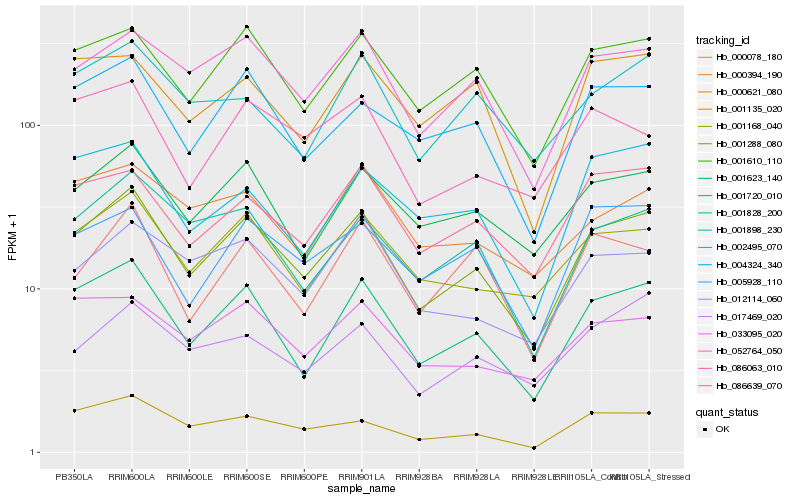
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001288_080 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 2 |
Hb_001720_010 |
0.0642809132 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001610_110 |
0.079122974 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 4 |
Hb_052764_050 |
0.081375756 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 5 |
Hb_001168_040 |
0.0883858449 |
- |
- |
- |
| 6 |
Hb_001623_140 |
0.0887355761 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 7 |
Hb_004324_340 |
0.094411404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001135_020 |
0.0948203907 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 9 |
Hb_086639_070 |
0.0952298446 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 10 |
Hb_001828_200 |
0.10061103 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 11 |
Hb_005928_110 |
0.1044089703 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 12 |
Hb_033095_020 |
0.1109627938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_017469_020 |
0.1109824994 |
- |
- |
- |
| 14 |
Hb_001898_230 |
0.111964877 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 15 |
Hb_012114_060 |
0.1124654443 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 16 |
Hb_000621_080 |
0.1131631764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002495_070 |
0.1146057474 |
- |
- |
PREDICTED: uncharacterized protein LOC105634731 [Jatropha curcas] |
| 18 |
Hb_000394_190 |
0.1154064343 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000078_180 |
0.1159692099 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 20 |
Hb_086063_010 |
0.1167066499 |
- |
- |
soluble diacylglycerol acyltransferase [Ricinus communis] |