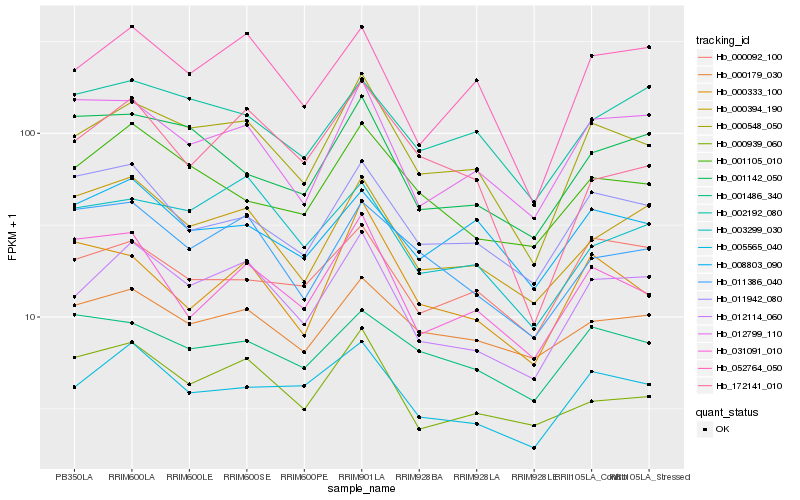
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000548_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 2 |
Hb_012114_060 |
0.0767542114 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 3 |
Hb_005565_040 |
0.0977146675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_052764_050 |
0.098157776 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 5 |
Hb_012799_110 |
0.104298845 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 6 |
Hb_011942_080 |
0.1050003545 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 7 |
Hb_000394_190 |
0.1056356121 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000179_030 |
0.1070321343 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001105_010 |
0.1081347421 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000333_100 |
0.1098445588 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002192_080 |
0.1110651703 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 12 |
Hb_003299_030 |
0.1110901701 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 13 |
Hb_011386_040 |
0.1111496987 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000092_100 |
0.1118165153 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 15 |
Hb_001486_340 |
0.1130502584 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 16 |
Hb_008803_090 |
0.1140935937 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |
| 17 |
Hb_000939_060 |
0.1147971439 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 18 |
Hb_031091_010 |
0.115943753 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 19 |
Hb_001142_050 |
0.1168316178 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 20 |
Hb_172141_010 |
0.1177964727 |
- |
- |
PREDICTED: uncharacterized protein LOC105641033 [Jatropha curcas] |