| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
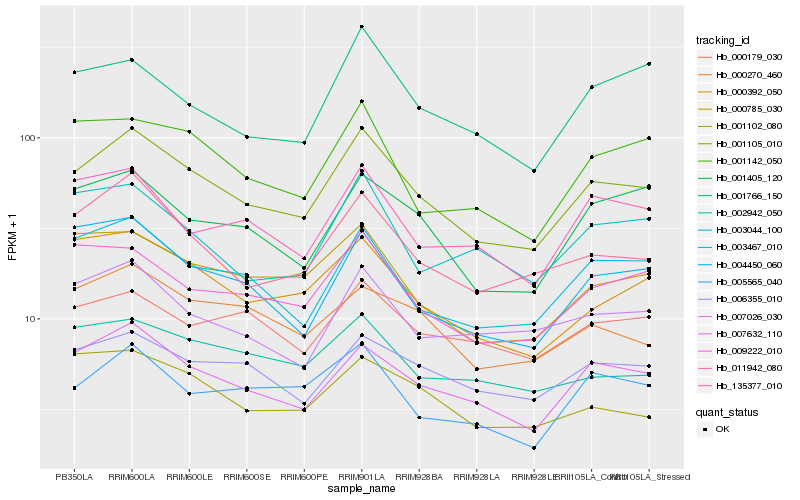
Hb_001105_010 |
0.0 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_007632_110 |
0.0660412314 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 3 |
Hb_135377_010 |
0.078952815 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 4 |
Hb_003467_010 |
0.0792106497 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 5 |
Hb_006355_010 |
0.086531902 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 6 |
Hb_009222_010 |
0.08725857 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_000392_050 |
0.0909211013 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000785_030 |
0.0910057452 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 9 |
Hb_001405_120 |
0.0930247047 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 10 |
Hb_005565_040 |
0.0931224635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007026_030 |
0.0946757696 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 12 |
Hb_011942_080 |
0.094692359 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 13 |
Hb_001142_050 |
0.0950640331 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 14 |
Hb_000270_460 |
0.0965686842 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 15 |
Hb_004450_060 |
0.0967480319 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000179_030 |
0.0978042244 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002942_050 |
0.0991326671 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001766_150 |
0.1008015942 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 19 |
Hb_001102_080 |
0.1013869169 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_003044_100 |
0.1018124185 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |