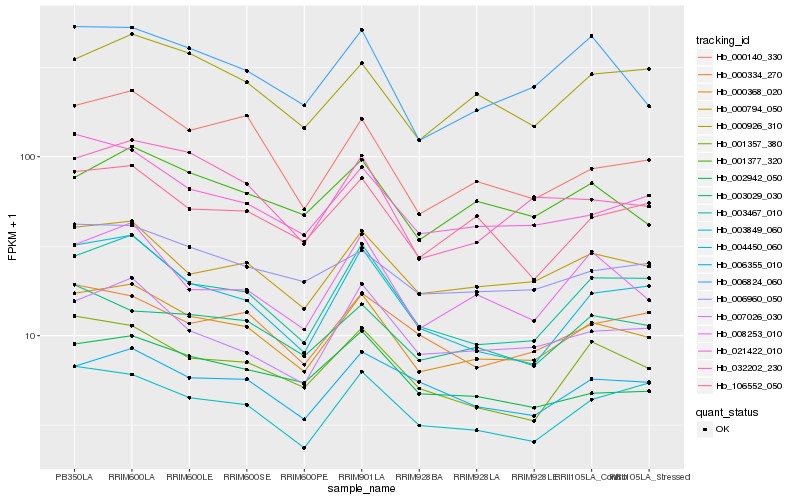
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000368_020 |
0.0 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000794_050 |
0.0551753136 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 3 |
Hb_003467_010 |
0.063183098 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_006960_050 |
0.0735116572 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 5 |
Hb_106552_050 |
0.0740192841 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 6 |
Hb_000926_310 |
0.0742732886 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 7 |
Hb_006824_060 |
0.0744576236 |
- |
- |
PREDICTED: uncharacterized protein LOC105650737 [Jatropha curcas] |
| 8 |
Hb_007026_030 |
0.0745694905 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 9 |
Hb_008253_010 |
0.074599939 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 10 |
Hb_001357_380 |
0.075337396 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_021422_010 |
0.0754354435 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001377_320 |
0.0761941784 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 13 |
Hb_032202_230 |
0.0767792843 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 14 |
Hb_003029_030 |
0.0771851733 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003849_060 |
0.0781203427 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000334_270 |
0.0809810026 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000140_330 |
0.0814118341 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 18 |
Hb_004450_060 |
0.0823972521 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002942_050 |
0.082639516 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 20 |
Hb_006355_010 |
0.0827865957 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |