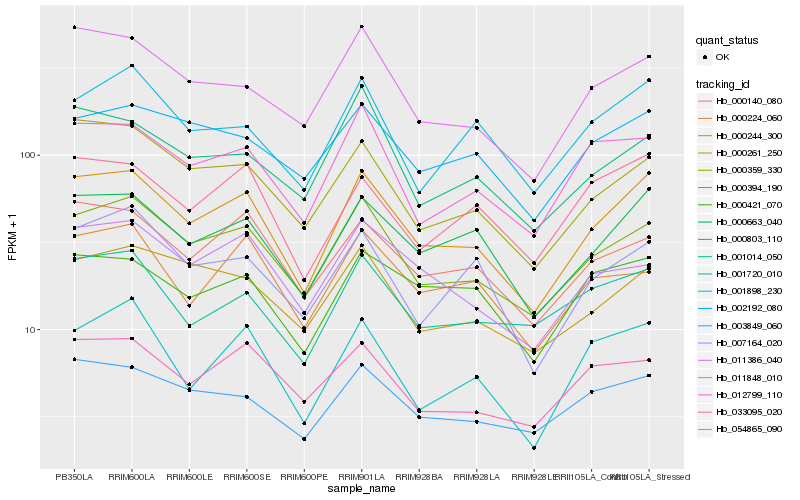
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000244_300 |
0.0 |
- |
- |
PREDICTED: probable 39S ribosomal protein L17, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000394_190 |
0.0633907211 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000663_040 |
0.0649738401 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000359_330 |
0.086140543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011848_010 |
0.0871834422 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 6 |
Hb_000140_080 |
0.0881168245 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003849_060 |
0.0893725066 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 8 |
Hb_012799_110 |
0.0923877137 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 9 |
Hb_054865_090 |
0.0941406645 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 10 |
Hb_000261_250 |
0.0945481661 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 11 |
Hb_000421_070 |
0.0961191886 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_033095_020 |
0.0974226831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002192_080 |
0.097645298 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 14 |
Hb_001720_010 |
0.0984232371 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_007164_020 |
0.0993728798 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 16 |
Hb_001014_050 |
0.0995255289 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 17 |
Hb_011386_040 |
0.0995454229 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001898_230 |
0.1009771488 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000224_060 |
0.1042726557 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 20 |
Hb_000803_110 |
0.1045619919 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |