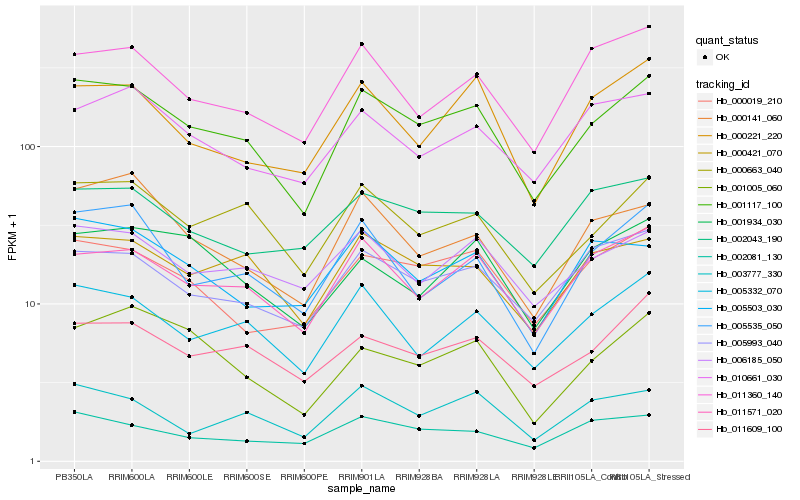
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_100 |
0.0 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 2 |
Hb_000663_040 |
0.0823154655 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011571_020 |
0.0855042543 |
- |
- |
PREDICTED: metal tolerance protein C4 [Jatropha curcas] |
| 4 |
Hb_005332_070 |
0.0896151867 |
- |
- |
PREDICTED: uncharacterized protein LOC105632487 [Jatropha curcas] |
| 5 |
Hb_000421_070 |
0.0911718737 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000019_210 |
0.0948724316 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 7 |
Hb_005993_040 |
0.0961525224 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit wdr4 [Jatropha curcas] |
| 8 |
Hb_011609_100 |
0.0970928717 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_006185_050 |
0.0996182135 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 10 |
Hb_000141_060 |
0.1012018035 |
- |
- |
PREDICTED: uncharacterized protein LOC105632248 [Jatropha curcas] |
| 11 |
Hb_002081_130 |
0.1039045213 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_005535_050 |
0.1042225423 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_003777_330 |
0.1057945619 |
- |
- |
ribonucleoside-diphosphate reductase large chain, putative [Ricinus communis] |
| 14 |
Hb_011360_140 |
0.106122418 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 15 |
Hb_005503_030 |
0.1071450439 |
- |
- |
PREDICTED: protein SCO1 homolog 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000221_220 |
0.1073788534 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 17 |
Hb_001005_060 |
0.107434864 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 18 |
Hb_010661_030 |
0.1089831254 |
- |
- |
hypothetical protein PHAVU_009G146200g [Phaseolus vulgaris] |
| 19 |
Hb_001934_030 |
0.1099185546 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 20 |
Hb_002043_190 |
0.11040528 |
- |
- |
rwd domain-containing protein, putative [Ricinus communis] |