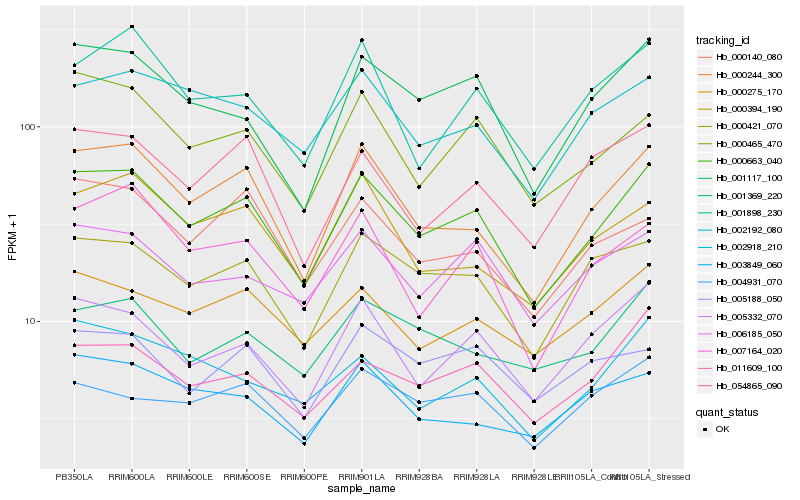
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000663_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000244_300 |
0.0649738401 |
- |
- |
PREDICTED: probable 39S ribosomal protein L17, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000421_070 |
0.0769940349 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_001117_100 |
0.0823154655 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 5 |
Hb_005332_070 |
0.0823583112 |
- |
- |
PREDICTED: uncharacterized protein LOC105632487 [Jatropha curcas] |
| 6 |
Hb_000465_470 |
0.0845446668 |
- |
- |
unknown [Picea sitchensis] |
| 7 |
Hb_011609_100 |
0.0876006901 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002192_080 |
0.0882289385 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 9 |
Hb_000275_170 |
0.08970595 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 10 |
Hb_006185_050 |
0.0899617683 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 11 |
Hb_007164_020 |
0.0906275171 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 12 |
Hb_000140_080 |
0.0932255073 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_054865_090 |
0.0932287028 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 14 |
Hb_000394_190 |
0.0937090214 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_001898_230 |
0.0941272311 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001369_220 |
0.0951348588 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002918_210 |
0.0964328811 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |
| 18 |
Hb_004931_070 |
0.099221877 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003849_060 |
0.09999906 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 20 |
Hb_005188_050 |
0.101592393 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 2B, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |