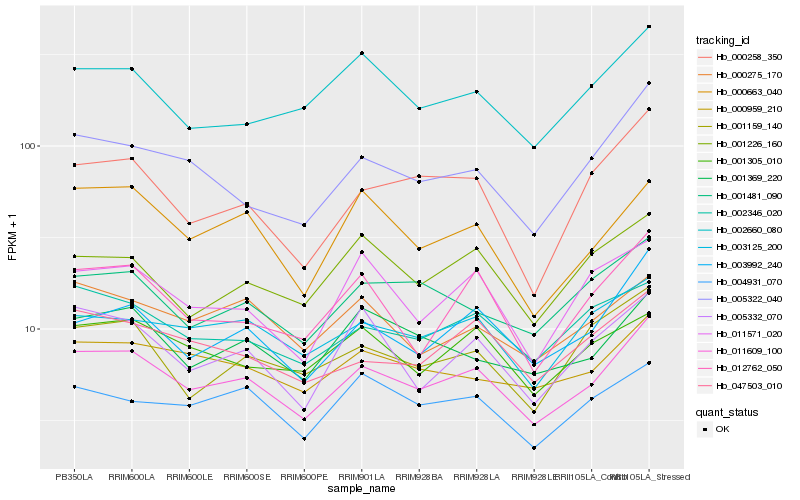
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_100 |
0.0 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_003992_240 |
0.0733688878 |
- |
- |
- |
| 3 |
Hb_005322_040 |
0.0822810103 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 4 |
Hb_003125_200 |
0.0831530206 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 5 |
Hb_047503_010 |
0.0842293587 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 6 |
Hb_000959_210 |
0.0848447288 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 7 |
Hb_002346_020 |
0.0852569073 |
- |
- |
- |
| 8 |
Hb_012762_050 |
0.0869740734 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 9 |
Hb_000663_040 |
0.0876006901 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001481_090 |
0.0885137891 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 11 |
Hb_005332_070 |
0.0900856144 |
- |
- |
PREDICTED: uncharacterized protein LOC105632487 [Jatropha curcas] |
| 12 |
Hb_001369_220 |
0.0901647354 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000258_350 |
0.0909434064 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 14 |
Hb_000275_170 |
0.091090275 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 15 |
Hb_001159_140 |
0.0917035517 |
- |
- |
hypothetical protein POPTR_0001s08090g [Populus trichocarpa] |
| 16 |
Hb_002660_080 |
0.0922461055 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 17 |
Hb_004931_070 |
0.0943037742 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001305_010 |
0.094571998 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011571_020 |
0.0950306936 |
- |
- |
PREDICTED: metal tolerance protein C4 [Jatropha curcas] |
| 20 |
Hb_001226_160 |
0.0950921159 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |