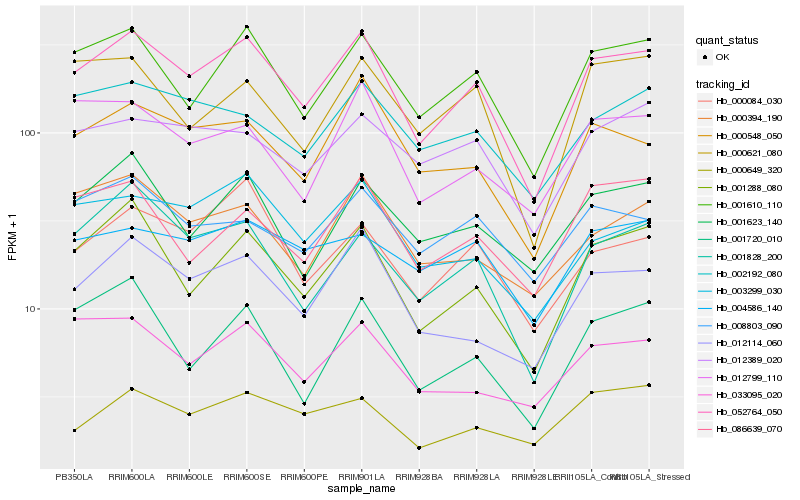
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052764_050 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 2 |
Hb_001610_110 |
0.0727034495 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 3 |
Hb_001288_080 |
0.081375756 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 4 |
Hb_012114_060 |
0.0916882736 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 5 |
Hb_000548_050 |
0.098157776 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 6 |
Hb_001828_200 |
0.0986400184 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 7 |
Hb_001623_140 |
0.1010041414 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 8 |
Hb_003299_030 |
0.1051851919 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000084_030 |
0.1061948944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002192_080 |
0.107725833 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 11 |
Hb_000394_190 |
0.1080102243 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_012389_020 |
0.1084889076 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 13 |
Hb_012799_110 |
0.1085658095 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 14 |
Hb_033095_020 |
0.1092335364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_086639_070 |
0.1108468439 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 16 |
Hb_000621_080 |
0.1110234525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001720_010 |
0.1117047197 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000649_320 |
0.1122707002 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004586_140 |
0.1130126476 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 20 |
Hb_008803_090 |
0.1132559082 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 6a [Jatropha curcas] |