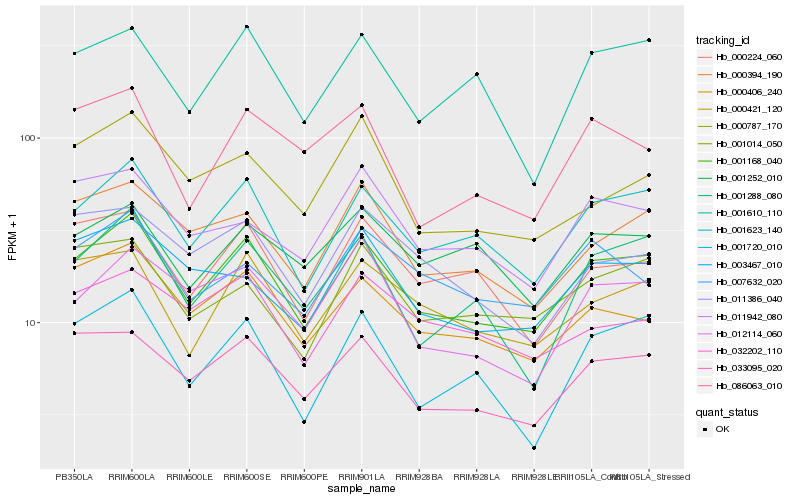
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001168_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_001623_140 |
0.0545156718 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 3 |
Hb_033095_020 |
0.0766102223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001288_080 |
0.0883858449 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 5 |
Hb_000421_120 |
0.0885024294 |
- |
- |
PREDICTED: NEDD8-specific protease 1 [Jatropha curcas] |
| 6 |
Hb_000406_240 |
0.0895788161 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 7 |
Hb_000394_190 |
0.0899215615 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001610_110 |
0.0955555897 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 9 |
Hb_000224_060 |
0.0958529653 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 10 |
Hb_001720_010 |
0.0961197904 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001014_050 |
0.0997521667 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 12 |
Hb_012114_060 |
0.1000802233 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 13 |
Hb_003467_010 |
0.1020995975 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_011942_080 |
0.1039471238 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 15 |
Hb_001252_010 |
0.106187397 |
- |
- |
hypothetical protein JCGZ_01389 [Jatropha curcas] |
| 16 |
Hb_000787_170 |
0.1063731978 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 17 |
Hb_007632_020 |
0.106548121 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 18 |
Hb_086063_010 |
0.1067972898 |
- |
- |
soluble diacylglycerol acyltransferase [Ricinus communis] |
| 19 |
Hb_032202_110 |
0.1072406869 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 20 |
Hb_011386_040 |
0.1078030093 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |