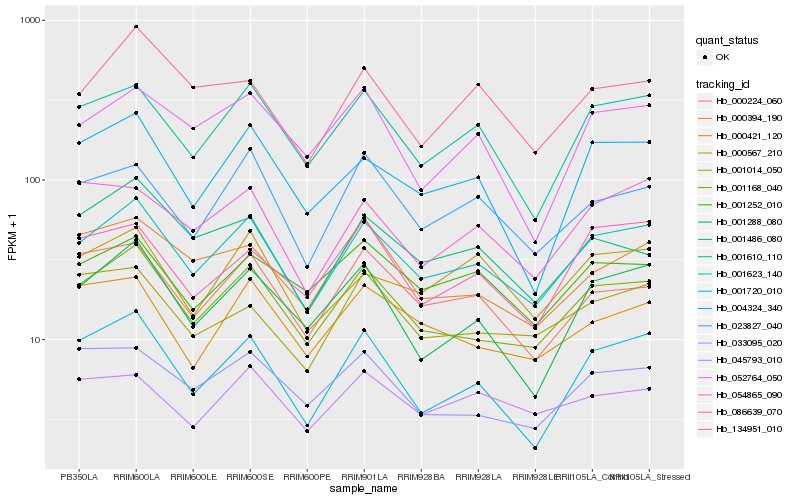
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_140 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 2 |
Hb_001168_040 |
0.0545156718 |
- |
- |
- |
| 3 |
Hb_001610_110 |
0.0703720446 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 4 |
Hb_001288_080 |
0.0887355761 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 5 |
Hb_001720_010 |
0.089536057 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_023827_040 |
0.0904175197 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 7 |
Hb_004324_340 |
0.091487447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001252_010 |
0.0935892277 |
- |
- |
hypothetical protein JCGZ_01389 [Jatropha curcas] |
| 9 |
Hb_000567_210 |
0.0941302955 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2-like [Jatropha curcas] |
| 10 |
Hb_000224_060 |
0.0945364091 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 11 |
Hb_054865_090 |
0.0977852646 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 12 |
Hb_000421_120 |
0.1007621476 |
- |
- |
PREDICTED: NEDD8-specific protease 1 [Jatropha curcas] |
| 13 |
Hb_052764_050 |
0.1010041414 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 14 |
Hb_086639_070 |
0.1010530589 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 15 |
Hb_045793_010 |
0.1018567408 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |
| 16 |
Hb_134951_010 |
0.1018968371 |
- |
- |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 17 |
Hb_033095_020 |
0.1022139384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000394_190 |
0.1028505685 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_001014_050 |
0.1041786159 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 20 |
Hb_001486_080 |
0.1068416812 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |