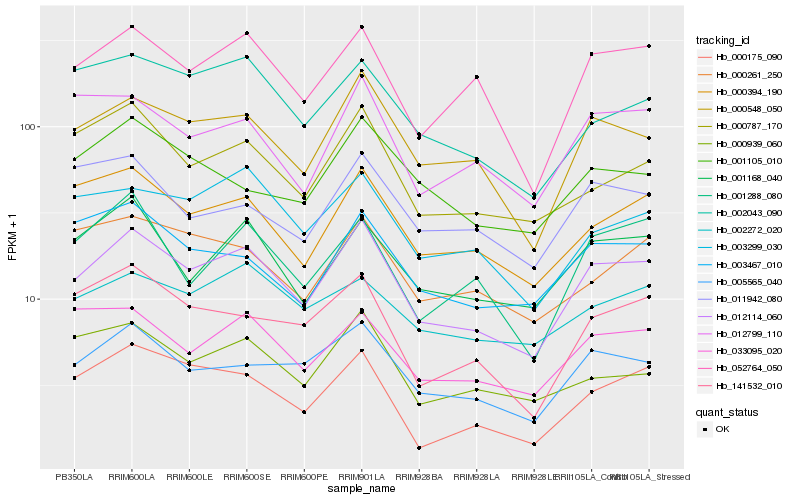
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012114_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 2 |
Hb_000548_050 |
0.0767542114 |
- |
- |
hypothetical protein POPTR_0008s12570g [Populus trichocarpa] |
| 3 |
Hb_005565_040 |
0.0878685375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_052764_050 |
0.0916882736 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 5 |
Hb_000394_190 |
0.0943769955 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001168_040 |
0.1000802233 |
- |
- |
- |
| 7 |
Hb_000787_170 |
0.1034341913 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 8 |
Hb_003299_030 |
0.107096995 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000175_090 |
0.1077610047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_033095_020 |
0.1081831712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_141532_010 |
0.1082882274 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 12 |
Hb_012799_110 |
0.1085178563 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 13 |
Hb_002043_090 |
0.1101370278 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 14 |
Hb_000939_060 |
0.1107737032 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 15 |
Hb_003467_010 |
0.1114365274 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_001288_080 |
0.1124654443 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 17 |
Hb_002272_020 |
0.1132691668 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 18 |
Hb_000261_250 |
0.1165948143 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 19 |
Hb_001105_010 |
0.117522288 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_011942_080 |
0.1197641382 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |