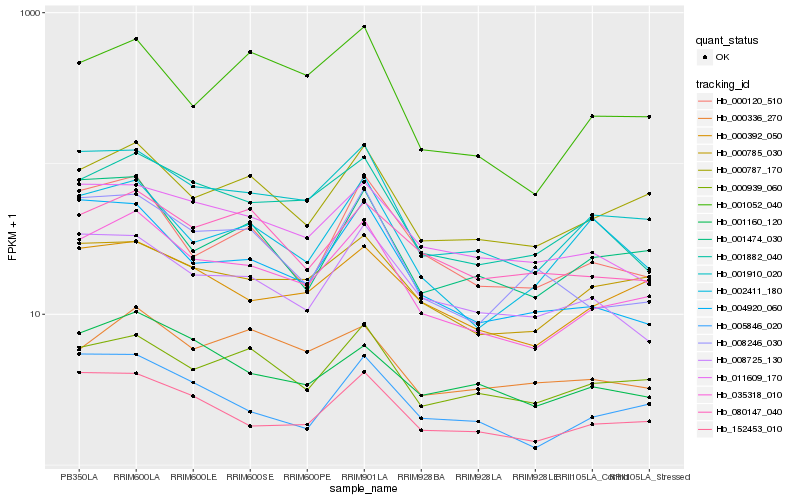
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035318_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 2 |
Hb_001910_020 |
0.0712206946 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 3 |
Hb_000787_170 |
0.0920178752 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 4 |
Hb_001882_040 |
0.0978368601 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 5 |
Hb_152453_010 |
0.10059761 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000392_050 |
0.1081097542 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000939_060 |
0.1083365753 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 8 |
Hb_000785_030 |
0.1108651002 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 9 |
Hb_004920_060 |
0.1109531252 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 10 |
Hb_000120_510 |
0.1127805505 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001160_120 |
0.115864155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001052_040 |
0.1169567929 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_011609_170 |
0.117374469 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 14 |
Hb_001474_030 |
0.1177509486 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 15 |
Hb_005846_020 |
0.1178000421 |
- |
- |
PREDICTED: tubulin-folding cofactor C [Jatropha curcas] |
| 16 |
Hb_008725_130 |
0.1200541088 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 17 |
Hb_080147_040 |
0.1202841412 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 18 |
Hb_008246_030 |
0.1208954185 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 19 |
Hb_002411_180 |
0.1221625043 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000336_270 |
0.1237130889 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |