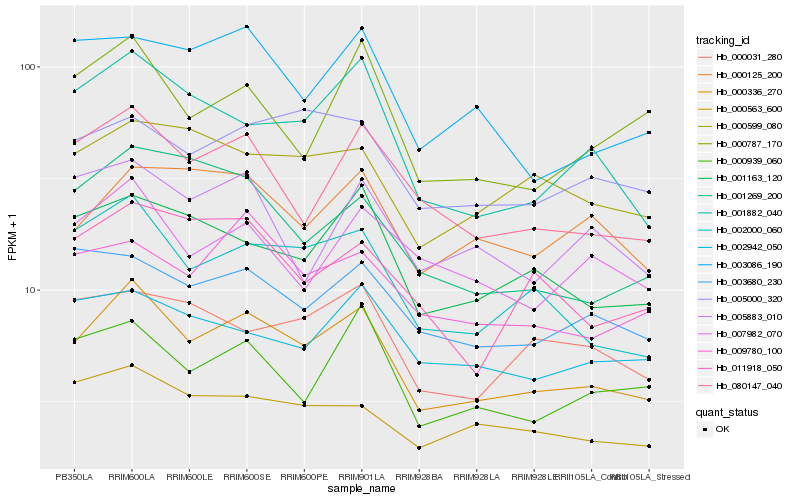
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_270 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002000_060 |
0.0878181677 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 3 |
Hb_080147_040 |
0.0962126543 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 4 |
Hb_001882_040 |
0.1014156629 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 5 |
Hb_001163_120 |
0.1033474489 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000563_600 |
0.1104679317 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 7 |
Hb_009780_100 |
0.1117156161 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 8 |
Hb_000939_060 |
0.1119979615 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 9 |
Hb_005000_320 |
0.113499294 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 10 |
Hb_003680_230 |
0.1139568221 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 11 |
Hb_003086_190 |
0.1145682268 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 12 |
Hb_000125_200 |
0.1147334371 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 13 |
Hb_000599_080 |
0.1156586796 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 14 |
Hb_000787_170 |
0.1173287892 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 15 |
Hb_001269_200 |
0.117607877 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 16 |
Hb_002942_050 |
0.1194969586 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_007982_070 |
0.1201603124 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 18 |
Hb_011918_050 |
0.1201851432 |
- |
- |
- |
| 19 |
Hb_000031_280 |
0.1220193736 |
- |
- |
- |
| 20 |
Hb_005883_010 |
0.1224186583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |