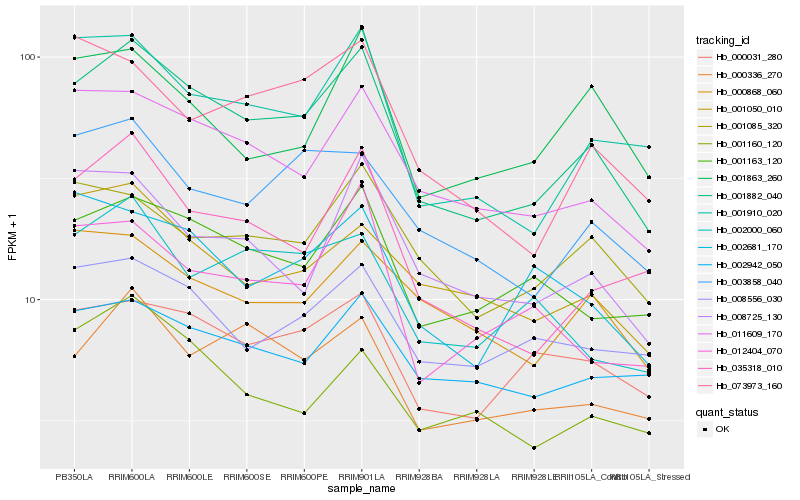
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001882_040 |
0.0 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 2 |
Hb_011609_170 |
0.0879494128 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 3 |
Hb_001910_020 |
0.0947030865 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_001163_120 |
0.0969679587 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_035318_010 |
0.0978368601 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 6 |
Hb_000031_280 |
0.1012081776 |
- |
- |
- |
| 7 |
Hb_000336_270 |
0.1014156629 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003858_040 |
0.1067735104 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 9 |
Hb_001085_320 |
0.1114015117 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_008556_030 |
0.1116182268 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002942_050 |
0.1131786798 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 12 |
Hb_008725_130 |
0.1134073246 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 13 |
Hb_001050_010 |
0.1143959884 |
- |
- |
Advillin [Gossypium arboreum] |
| 14 |
Hb_073973_160 |
0.1164432889 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 15 |
Hb_000868_060 |
0.1165497773 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_001160_120 |
0.1165991004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002681_170 |
0.11744588 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 18 |
Hb_002000_060 |
0.1180707373 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 19 |
Hb_012404_070 |
0.1182238259 |
- |
- |
PREDICTED: uncharacterized protein LOC105629465 [Jatropha curcas] |
| 20 |
Hb_001863_260 |
0.1186315868 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |