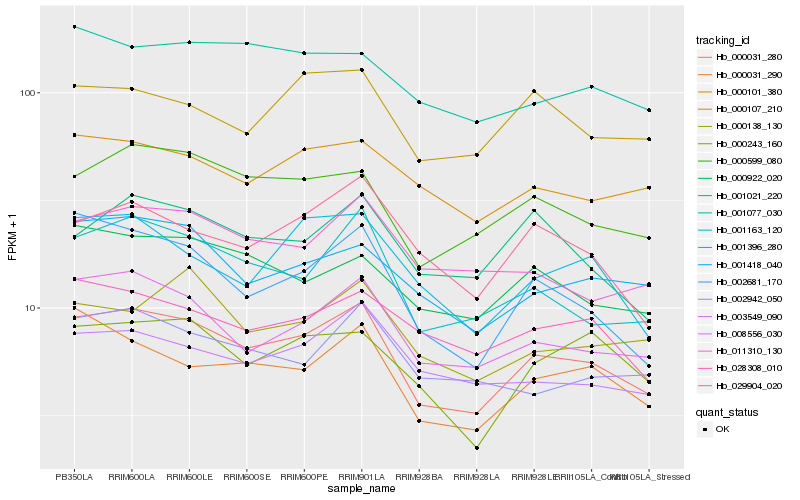
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_280 |
0.0 |
- |
- |
- |
| 2 |
Hb_008556_030 |
0.0740867563 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001163_120 |
0.0745924842 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000599_080 |
0.0780362881 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 5 |
Hb_002681_170 |
0.0818663794 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 6 |
Hb_000031_290 |
0.0868649994 |
- |
- |
hypothetical protein POPTR_0006s28540g [Populus trichocarpa] |
| 7 |
Hb_001418_040 |
0.0902588612 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 8 |
Hb_000922_020 |
0.0930284902 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_001396_280 |
0.0939665759 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 10 |
Hb_000243_160 |
0.095088826 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 11 |
Hb_003549_090 |
0.0958835392 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 12 |
Hb_029904_020 |
0.0961962794 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 13 |
Hb_011310_130 |
0.0981501866 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000101_380 |
0.0988238357 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 15 |
Hb_002942_050 |
0.0990142743 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 16 |
Hb_028308_010 |
0.0993931191 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000107_210 |
0.0995560776 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 18 |
Hb_001021_220 |
0.0998372028 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001077_030 |
0.1002455241 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000138_130 |
0.1008534107 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |