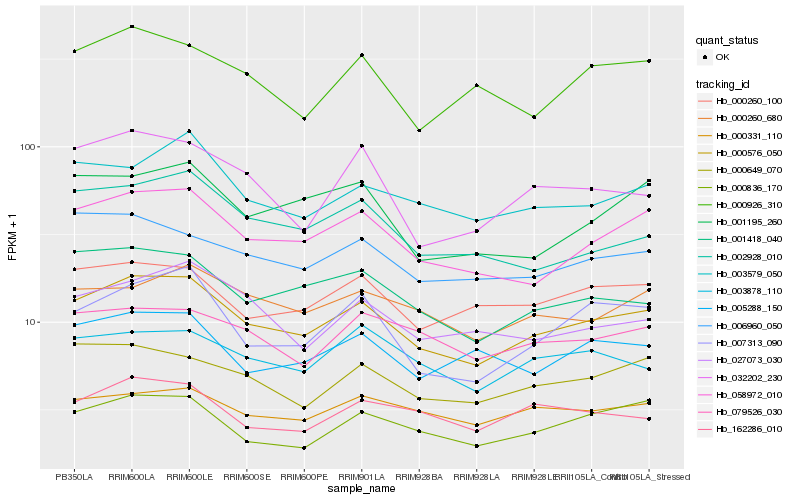
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000576_050 |
0.0 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 2 |
Hb_058972_010 |
0.0592326834 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001418_040 |
0.0721539549 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 4 |
Hb_000649_070 |
0.0772659607 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 5 |
Hb_007313_090 |
0.078616343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006960_050 |
0.0790483449 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 7 |
Hb_000260_100 |
0.0793847498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003579_050 |
0.0803931214 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 9 |
Hb_000836_170 |
0.081149652 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 10 |
Hb_000260_680 |
0.0828678226 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 11 |
Hb_027073_030 |
0.0845024903 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 12 |
Hb_005288_150 |
0.0855449802 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_002928_010 |
0.0856415441 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 14 |
Hb_003878_110 |
0.085887719 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 15 |
Hb_000926_310 |
0.0870862302 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 16 |
Hb_001195_260 |
0.0872526016 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 17 |
Hb_079526_030 |
0.0875340517 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 18 |
Hb_032202_230 |
0.088607034 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 19 |
Hb_162286_010 |
0.0886599954 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 20 |
Hb_000331_110 |
0.089684839 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |