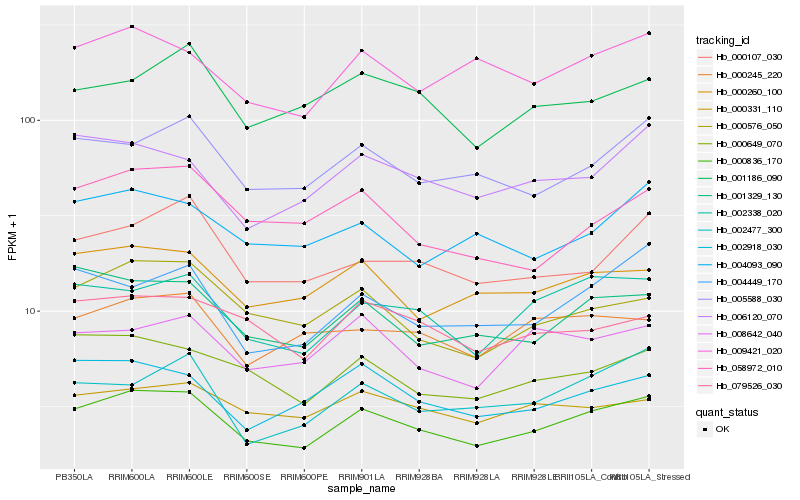
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_170 |
0.0 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 2 |
Hb_002338_020 |
0.0755021124 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 3 |
Hb_005588_030 |
0.0804169147 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 4 |
Hb_000576_050 |
0.081149652 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 5 |
Hb_001329_130 |
0.0821352764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000649_070 |
0.0830166436 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 7 |
Hb_000260_100 |
0.084430833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000107_030 |
0.0854603762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009421_020 |
0.0875714429 |
- |
- |
PREDICTED: caltractin-like [Jatropha curcas] |
| 10 |
Hb_002918_030 |
0.0878568224 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 11 |
Hb_004449_170 |
0.0887375499 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006120_070 |
0.0899361122 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 13 |
Hb_004093_090 |
0.0936239942 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 14 |
Hb_000331_110 |
0.0936452801 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_001186_090 |
0.0937888833 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 16 |
Hb_000245_220 |
0.0938199367 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 17 |
Hb_079526_030 |
0.0938291096 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 18 |
Hb_008642_040 |
0.093934234 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_058972_010 |
0.0945833781 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002477_300 |
0.0956080573 |
- |
- |
transcription factor, putative [Ricinus communis] |