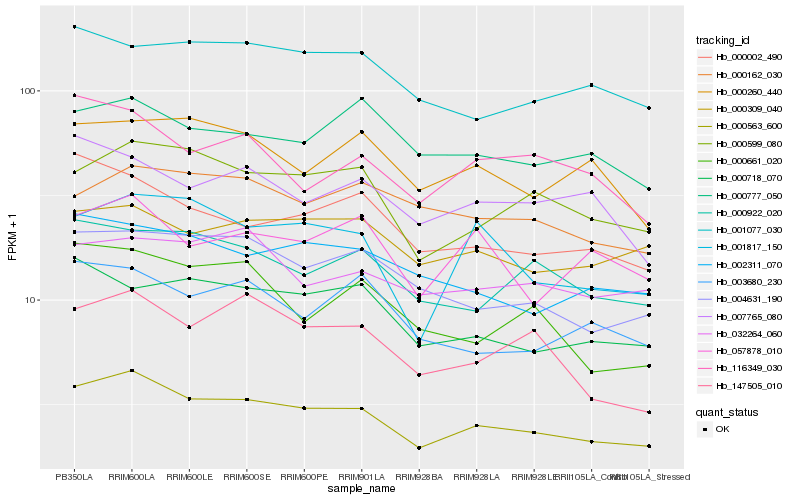
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_600 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 2 |
Hb_000599_080 |
0.0762021898 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 3 |
Hb_004631_190 |
0.0782799843 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 4 |
Hb_002311_070 |
0.0860361099 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 5 |
Hb_000922_020 |
0.0869314994 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000002_490 |
0.0878546132 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 7 |
Hb_000718_070 |
0.0881519606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001817_150 |
0.0891823409 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_147505_010 |
0.0902325759 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 10 |
Hb_000309_040 |
0.0922965048 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 11 |
Hb_001077_030 |
0.0932096116 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003680_230 |
0.0932769981 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 13 |
Hb_000260_440 |
0.0938754587 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 14 |
Hb_032264_060 |
0.0956632566 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 15 |
Hb_000777_050 |
0.0961230599 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 16 |
Hb_000162_030 |
0.0975345581 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_057878_010 |
0.0975701441 |
- |
- |
hypothetical protein VITISV_036304 [Vitis vinifera] |
| 18 |
Hb_007765_080 |
0.0978130663 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 19 |
Hb_116349_030 |
0.0998473875 |
- |
- |
PREDICTED: zinc finger MYND domain-containing protein 15 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000661_020 |
0.0999208072 |
- |
- |
BnaC08g27190D [Brassica napus] |