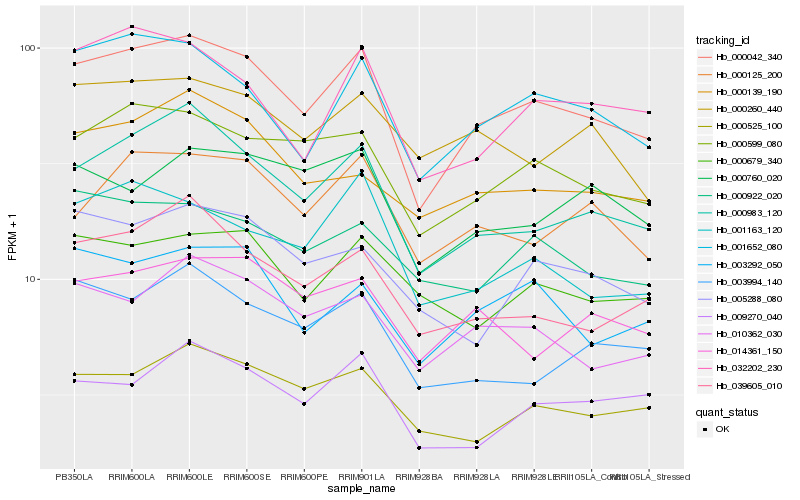
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_340 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 2 |
Hb_000983_120 |
0.0728229822 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000599_080 |
0.0740702799 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 4 |
Hb_001652_080 |
0.0775694353 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 5 |
Hb_010362_030 |
0.0861829656 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_000525_100 |
0.0865614031 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 7 |
Hb_003292_050 |
0.0889231669 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 8 |
Hb_032202_230 |
0.0899769126 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 9 |
Hb_000125_200 |
0.0918714229 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 10 |
Hb_039605_010 |
0.0938150844 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001163_120 |
0.0941919876 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_014361_150 |
0.0951422984 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 13 |
Hb_009270_040 |
0.0952709313 |
- |
- |
- |
| 14 |
Hb_000922_020 |
0.0963853196 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000679_340 |
0.0995523095 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000139_190 |
0.099719564 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003994_140 |
0.0997637811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005288_080 |
0.0998334018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000260_440 |
0.1015349404 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 20 |
Hb_000760_020 |
0.1016514992 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |