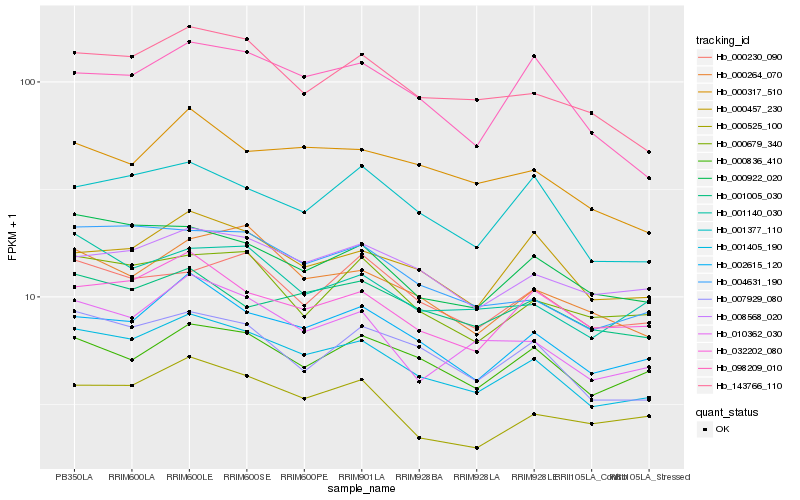
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_190 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002615_120 |
0.055365448 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_143766_110 |
0.0588834169 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 4 |
Hb_007929_080 |
0.06026951 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 5 |
Hb_004631_190 |
0.0608862336 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 6 |
Hb_000922_020 |
0.0641416498 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_010362_030 |
0.0649135387 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_000264_070 |
0.0689213969 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 9 |
Hb_008568_020 |
0.0689484544 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 10 |
Hb_032202_080 |
0.0699383815 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001377_110 |
0.0702152447 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 12 |
Hb_001140_030 |
0.0712786685 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 13 |
Hb_000317_510 |
0.0719404281 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 14 |
Hb_000457_230 |
0.0722464981 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000679_340 |
0.0739542165 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000836_410 |
0.0740297731 |
- |
- |
sec10, putative [Ricinus communis] |
| 17 |
Hb_000230_090 |
0.0749718962 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000525_100 |
0.0755779308 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 19 |
Hb_098209_010 |
0.0764648839 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 20 |
Hb_001005_030 |
0.077197287 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |