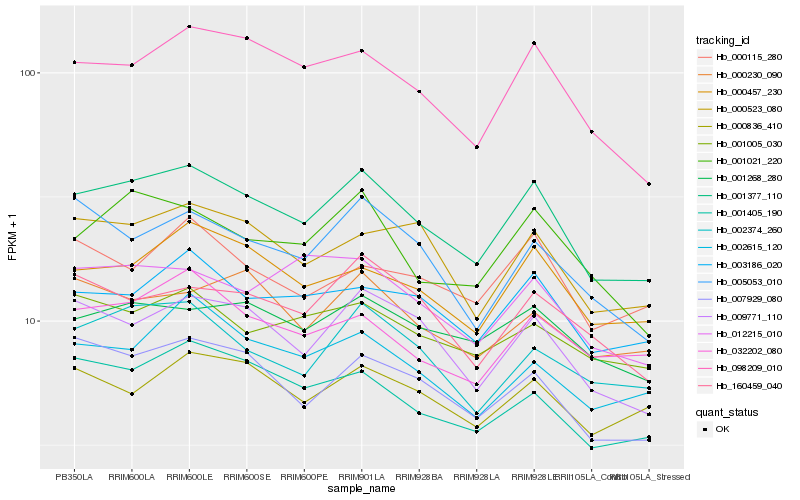
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_110 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 2 |
Hb_098209_010 |
0.0629997191 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 3 |
Hb_009771_110 |
0.0631729577 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 4 |
Hb_000457_230 |
0.0647625157 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007929_080 |
0.0649091769 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 6 |
Hb_003186_020 |
0.0677887159 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001021_220 |
0.069054735 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001405_190 |
0.0702152447 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001268_280 |
0.0705704082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000523_080 |
0.0707867595 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 11 |
Hb_002374_260 |
0.0712572246 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 12 |
Hb_032202_080 |
0.0770424518 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002615_120 |
0.0773310738 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_160459_040 |
0.0774315475 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 15 |
Hb_000836_410 |
0.0789584141 |
- |
- |
sec10, putative [Ricinus communis] |
| 16 |
Hb_001005_030 |
0.0792137636 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 17 |
Hb_012215_010 |
0.0798935259 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 18 |
Hb_005053_010 |
0.0801054393 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 19 |
Hb_000115_280 |
0.0810238695 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 20 |
Hb_000230_090 |
0.0814519339 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |