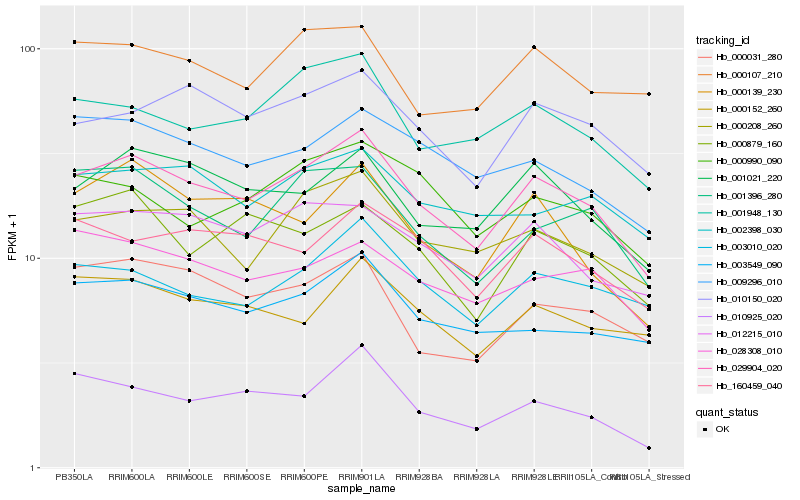
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029904_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 2 |
Hb_001021_220 |
0.079950613 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000208_260 |
0.0807765475 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_012215_010 |
0.0827332853 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 5 |
Hb_010150_020 |
0.0839222014 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 6 |
Hb_001948_130 |
0.0852994988 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 7 |
Hb_003549_090 |
0.0868962253 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 8 |
Hb_009296_010 |
0.0880624548 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 9 |
Hb_001396_280 |
0.0889234924 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 10 |
Hb_160459_040 |
0.0910742234 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 11 |
Hb_003010_020 |
0.0912267955 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_002398_030 |
0.0938964256 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000152_260 |
0.0945221062 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 14 |
Hb_028308_010 |
0.09536235 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000031_280 |
0.0961962794 |
- |
- |
- |
| 16 |
Hb_010925_020 |
0.096292515 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 17 |
Hb_000139_230 |
0.096464857 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 18 |
Hb_000990_090 |
0.0970833337 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_000107_210 |
0.0981234841 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_000879_160 |
0.0982617608 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |