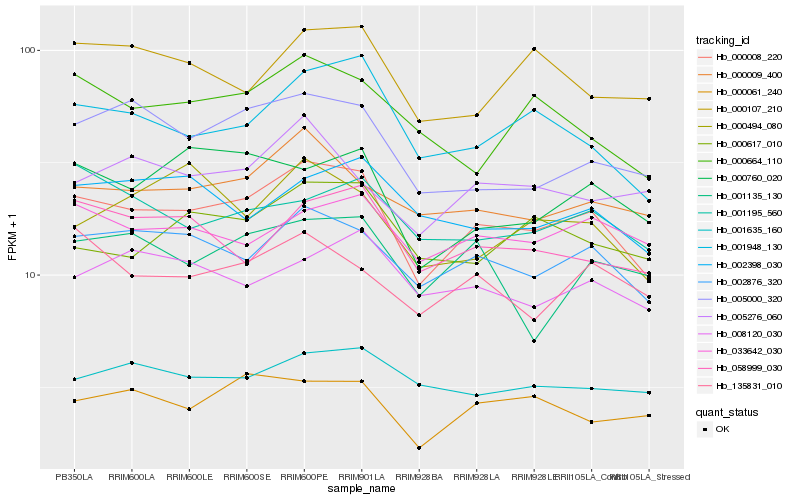
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_220 |
0.0 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 10 [Jatropha curcas] |
| 2 |
Hb_002876_320 |
0.0721058951 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 3 |
Hb_001948_130 |
0.0855150415 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 4 |
Hb_000009_400 |
0.0948894664 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 5 |
Hb_005000_320 |
0.0957842281 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 6 |
Hb_058999_030 |
0.0986728555 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000760_020 |
0.1001661148 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_033642_030 |
0.1002533284 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000061_240 |
0.1009898516 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_135831_010 |
0.1029682692 |
- |
- |
hypothetical protein JCGZ_13374 [Jatropha curcas] |
| 11 |
Hb_000664_110 |
0.1031230195 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 12 |
Hb_002398_030 |
0.1051723645 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000617_010 |
0.1052149084 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 14 |
Hb_001635_160 |
0.1053844878 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 15 |
Hb_008120_030 |
0.1057804339 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 16 |
Hb_000107_210 |
0.107734636 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_001195_560 |
0.1079506498 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005276_060 |
0.1085328125 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000494_080 |
0.1091808112 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001135_130 |
0.1111543899 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |