| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
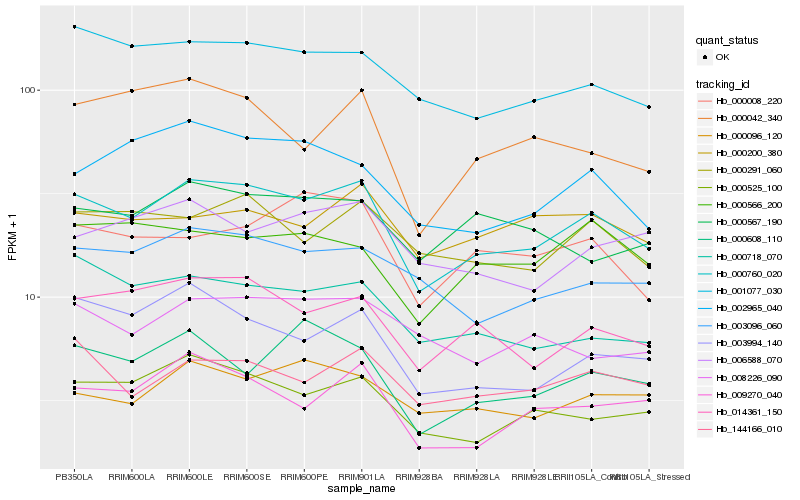
Hb_000760_020 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_014361_150 |
0.0820093111 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 3 |
Hb_001077_030 |
0.0862603678 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000525_100 |
0.086924603 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 5 |
Hb_144166_010 |
0.0912623856 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 6 |
Hb_000291_060 |
0.0918202723 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 7 |
Hb_003096_060 |
0.0921834943 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 8 |
Hb_009270_040 |
0.0947786254 |
- |
- |
- |
| 9 |
Hb_000096_120 |
0.095589872 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 10 |
Hb_003994_140 |
0.0969184319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002965_040 |
0.0978139337 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 12 |
Hb_000718_070 |
0.0982577519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000008_220 |
0.1001661148 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 10 [Jatropha curcas] |
| 14 |
Hb_008226_090 |
0.101609571 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000042_340 |
0.1016514992 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 16 |
Hb_000566_200 |
0.1020199725 |
- |
- |
PREDICTED: uncharacterized protein LOC105644834 [Jatropha curcas] |
| 17 |
Hb_000608_110 |
0.1025232881 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 18 |
Hb_006588_070 |
0.1034752483 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 19 |
Hb_000567_190 |
0.1037485862 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000200_380 |
0.1046020563 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |